| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,001,656 – 2,001,752 |
| Length | 96 |
| Max. P | 0.753623 |

| Location | 2,001,656 – 2,001,752 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.40 |
| Shannon entropy | 0.47107 |
| G+C content | 0.38434 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -13.11 |
| Energy contribution | -13.36 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
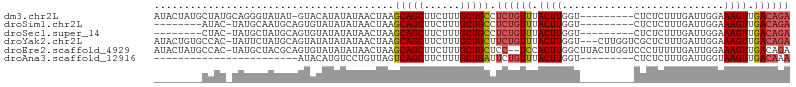
>dm3.chr2L 2001656 96 + 23011544 AUACUAUGCUAUGCAGGGUAUAU-GUACAUAUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUUACUUGGU---------CUCUCUUUGAUUGGAAAGUUGACAGA .(((((((((......)))))).-))).............(((((......))))).((((((.((((.((---------(.......)))....)))).)))))) ( -23.40, z-score = -1.08, R) >droSim1.chr2L 1970157 88 + 22036055 --------AUAC-UAUGCAAUGCAGUGUAUAUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUUACUUGGU---------CUCUCUUUGAUUGGAAAGUUGACAGA --------....-((((((......)))))).........(((((......))))).((((((.((((.((---------(.......)))....)))).)))))) ( -21.20, z-score = -1.19, R) >droSec1.super_14 1948515 88 + 2068291 --------CUAC-UAUGCUAUGCAGUGUAUAUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUUACUUGGU---------CUCUCUUUGAUUGGAAAGUUGACAGA --------....-(((((........))))).........(((((......))))).((((((.((((.((---------(.......)))....)))).)))))) ( -20.60, z-score = -1.00, R) >droYak2.chr2L 1986585 102 + 22324452 AUACUGUGCCAC-UAUUCUAUGCAGUAUAUAUAUAACUAAGCAGCUUCUUUGCUGCUUCUGUUUACUUGGU---CUUGGUCGCUCUUUGAUUGGAAAGUUGACAGA (((((((.....-........)))))))...........((((((......))))))((((((.((((...---.(..((((.....))))..).)))).)))))) ( -27.42, z-score = -2.18, R) >droEre2.scaffold_4929 2047531 103 + 26641161 AUACUAUGCCAC-UAUGCUACGCAGUGUAUAUAUAACUAAGCAGCUUCUUUGCUGCUCC--UCCACUUGGCUUACUUGGUCCCUUUUUGAUUGGAAAGUUGACAGA .......((((.-(((((........)))))........((((((......))))))..--......)))).......(((.((((((....))))))..)))... ( -22.60, z-score = -0.64, R) >droAna3.scaffold_12916 9716688 73 - 16180835 ------------------------AUACAUGUCCUGUUAGUCAGCUUCUUUGCUGAUUCUGUUUACUUGGU---------CUCUCUUUGAUUGGUAAGUUGACAAA ------------------------.....((((.....(((((((......)))))))...((((((.(((---------(.......))))))))))..)))).. ( -15.60, z-score = -1.69, R) >consensus ________CUAC_UAUGCUAUGCAGUAUAUAUAUAACUAAGCAGCUUCUUUGCUGCCUCUGUUUACUUGGU_________CUCUCUUUGAUUGGAAAGUUGACAGA ........................................(((((......))))).((((((.((((...........................)))).)))))) (-13.11 = -13.36 + 0.25)

| Location | 2,001,656 – 2,001,752 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.40 |
| Shannon entropy | 0.47107 |
| G+C content | 0.38434 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -11.88 |
| Energy contribution | -11.66 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2001656 96 - 23011544 UCUGUCAACUUUCCAAUCAAAGAGAG---------ACCAAGUAAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAUAUGUAC-AUAUACCCUGCAUAGCAUAGUAU (((((..((((.....((.....)).---------...))))..)))))((((((......))))))...((((.((((((.-........))))))..))))... ( -19.20, z-score = -1.28, R) >droSim1.chr2L 1970157 88 - 22036055 UCUGUCAACUUUCCAAUCAAAGAGAG---------ACCAAGUAAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAUAUACACUGCAUUGCAUA-GUAU-------- (((((..((((.....((.....)).---------...))))..)))))((((((......)))))).........((((..(((...)))..-))))-------- ( -18.00, z-score = -1.11, R) >droSec1.super_14 1948515 88 - 2068291 UCUGUCAACUUUCCAAUCAAAGAGAG---------ACCAAGUAAACAGAGGCAGCAAAGAAGCUGCUUAGUUAUAUAUACACUGCAUAGCAUA-GUAG-------- (((((..((((.....((.....)).---------...))))..)))))((((((......))))))..............((((........-))))-------- ( -18.20, z-score = -1.34, R) >droYak2.chr2L 1986585 102 - 22324452 UCUGUCAACUUUCCAAUCAAAGAGCGACCAAG---ACCAAGUAAACAGAAGCAGCAAAGAAGCUGCUUAGUUAUAUAUAUACUGCAUAGAAUA-GUGGCACAGUAU .((((..((((((...((.......))....)---)..))))..))))(((((((......)))))))..........(((((((((......-)))...)))))) ( -18.30, z-score = -0.50, R) >droEre2.scaffold_4929 2047531 103 - 26641161 UCUGUCAACUUUCCAAUCAAAAAGGGACCAAGUAAGCCAAGUGGA--GGAGCAGCAAAGAAGCUGCUUAGUUAUAUAUACACUGCGUAGCAUA-GUGGCAUAGUAU .((((..(((((((..........)))..))))..((((.((((.--.(((((((......)))))))..))))...(((.....))).....-.))))))))... ( -23.00, z-score = -0.26, R) >droAna3.scaffold_12916 9716688 73 + 16180835 UUUGUCAACUUACCAAUCAAAGAGAG---------ACCAAGUAAACAGAAUCAGCAAAGAAGCUGACUAACAGGACAUGUAU------------------------ ..((((.((((.....((.....)).---------...))))........(((((......))))).......)))).....------------------------ ( -11.30, z-score = -1.22, R) >consensus UCUGUCAACUUUCCAAUCAAAGAGAG_________ACCAAGUAAACAGAAGCAGCAAAGAAGCUGCUUAGUUAUAUAUACACUGCAUAGCAUA_GUAG________ .((((..((((...........................))))..))))(((((((......)))))))...................................... (-11.88 = -11.66 + -0.22)

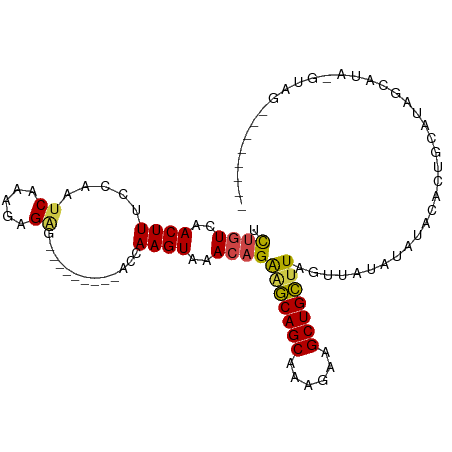
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:48 2011