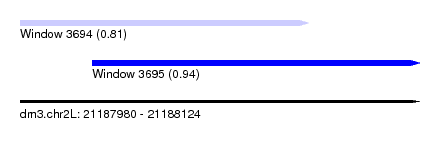
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,187,980 – 21,188,124 |
| Length | 144 |
| Max. P | 0.936789 |

| Location | 21,187,980 – 21,188,084 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.55 |
| Shannon entropy | 0.50381 |
| G+C content | 0.57112 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.36 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
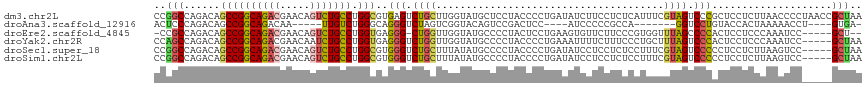
>dm3.chr2L 21187980 104 + 23011544 GAAUUCUAUAAGCCCGCACCCAGUGACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGAGUCUGCUUGGUAUGCUCCUACCCCUGAUAUCUUCCUC ..........(((.(((.....)))(((((.(((((.((((((((((.....))))))).)))....))))).)))))..)))..................... ( -33.70, z-score = -1.92, R) >droPer1.super_5 326678 84 + 6813705 GAAUUCAAUAAGCCCGCACCCAGAGUCAGACCAGACAGCCGGCAGACGA-----UUGUCUGGGCCAGGUCUGGUUC---UGUCUCUGUCUCU------------ ....................(((((.((((((((((.(((..(((((..-----..))))))))...)))))).))---)).))))).....------------ ( -34.00, z-score = -2.96, R) >dp4.chr4_group1 388305 84 - 5278887 GAAUUCAAUAAGCCCGCACCCAGAGUCAGACCAGACAGCCGGCAGACGA-----UUGUCUGGGCCAGGUCUGGUUC---UGGCUCUGUCUCU------------ ....................((((((((((((((((.(((..(((((..-----..))))))))...)))))).))---)))))))).....------------ ( -37.30, z-score = -3.49, R) >droAna3.scaffold_12916 11680037 95 - 16180835 GGAUUCAAUAAGCCCGCACCCAGUGAACUCCCAGACAGCCGGCAGACAA-----UUGUCUGGGCAGGGUCUAGUCGGUACAGUCCGACUCC----AUCCCCCGC (((.....((..(((.(((...)))....(((((((((..(.....)..-----)))))))))..)))..))(((((......))))))))----......... ( -31.00, z-score = -1.51, R) >droEre2.scaffold_4845 19279135 102 - 22589142 GAAUUCAAUAAGCCCGCACCCAGUGACCG-CCAGACAGCCGGCAGACGAACAGUCUGCCUGGUGAGGG-CUGGUUGGUAUGCCCCUACUCCUGAAGUGUUCUUC ....(((((.(((((.((((........(-(......)).(((((((.....))))))).)))).)))-)).)))))...............((((....)))) ( -35.50, z-score = -1.58, R) >droYak2.chr2R 19553451 104 - 21139217 GAAUUCAAUAAACCCGCACCCAGUGACCAGCCAGACAGCCGGCAGACGAACAAUCUGCCUGGUGAGGGUCUGGUUGGUAUGCCCCUACCCCUGAAAUUUUCUUU ...((((.......(((.....)))(((((((((((.(((((((((.......)))))).)))....))))))))))).............))))......... ( -36.50, z-score = -3.37, R) >droSec1.super_18 1109096 104 + 1342155 GAAUUCUAUAAGCCCGCACCCAGUUACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAUCCUCCUC ...........((((((.((........(((......)))(((((((.....))))))).)).))))))..((.......))...................... ( -31.00, z-score = -1.58, R) >droSim1.chr2L 20771009 104 + 22036055 GAAUUCUAUAAGCCCGCACCCAGUUACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAUCCUCCUC ...........((((((.((........(((......)))(((((((.....))))))).)).))))))..((.......))...................... ( -31.00, z-score = -1.58, R) >consensus GAAUUCAAUAAGCCCGCACCCAGUGACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGAGGGUCUGGUUGGUAUGCCCCUACCCCUGA_AUCCUCCUC ...............................(((((..(((((((.........))))).)).....)))))................................ (-12.58 = -12.36 + -0.22)
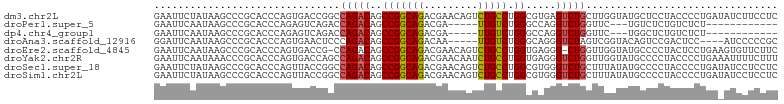
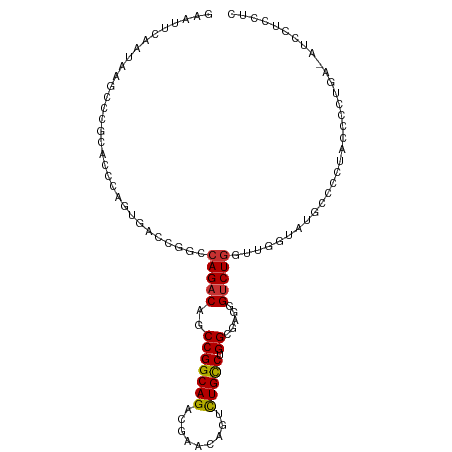
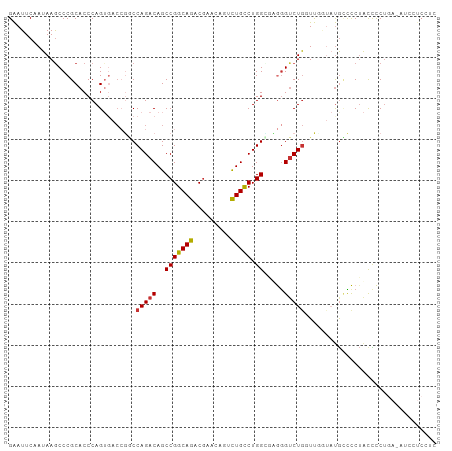
| Location | 21,188,006 – 21,188,124 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.24 |
| Shannon entropy | 0.50449 |
| G+C content | 0.57791 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
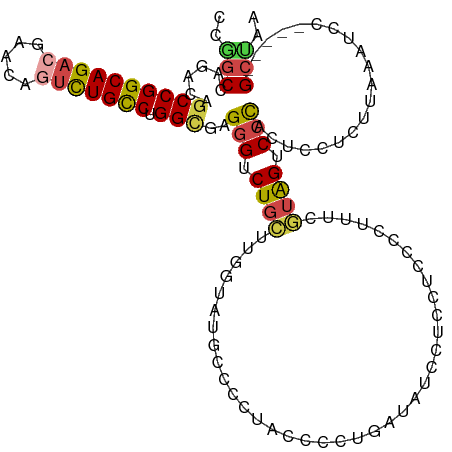
>dm3.chr2L 21188006 118 + 23011544 CCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGAGUCUGCUUGGUAUGCUCCUACCCCUGAUAUCUUCCUCUCAUUUCGUAGUCCCGCUCCUCUUAACCCCUAACCGCUAA .(((..((...((((((((((.....))))))).)))(((.(.((((..((((......))))...((.....))..........)))).).)))............))..))).... ( -32.20, z-score = -1.62, R) >droAna3.scaffold_12916 11680063 97 - 16180835 ACUCCCAGACAGCCGGCAGACAA-----UUGUCUGGGCAGGGUCUAGUCGGUACAGUCCGACUCC----AUCCCCCGCCA-------GGUCCUGUACCACUAAAAACCU----GUGA- ....((((((((..(.....)..-----))))))))(((((...((((.(((((((...((((..----...........-------)))))))))))))))....)))----))..- ( -33.42, z-score = -1.63, R) >droEre2.scaffold_4845 19279161 109 - 22589142 -CCGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGUGAGGG-CUGGUUGGUAUGCCCCUACUCCUGAAGUGUUCUUCCCGUGGUUUAGCCCCACUCCUCCCAAAUCC-----GCU-- -.........(((.(((((((.....)))))))(((.((((.-.(((..(((..(((...((....((((....))))..)))))...))))))..))))))).....-----)))-- ( -36.40, z-score = -0.90, R) >droYak2.chr2R 19553477 113 - 21139217 CCAGCCAGACAGCCGGCAGACGAACAAUCUGCCUGGUGAGGGUCUGGUUGGUAUGCCCCUACCCCUGAAAUUUUCUUUCCCUGCUUUAGUCCCACUCCUCCCAAAUCC-----GCUAA ((((((((((.(((((((((.......)))))).)))....)))))))))).........................................................-----..... ( -31.30, z-score = -1.16, R) >droSec1.super_18 1109122 113 + 1342155 CCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAUCCUCCUCUCCUUUCGUAGUCCCCCUCCUCUUAAGUCC-----GCUAA ..(((..(((.((((((((((.....))))))).)))(.(((.((((......................................)))).))).).........))).-----))).. ( -32.36, z-score = -1.92, R) >droSim1.chr2L 20771035 113 + 22036055 CCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAUCCUCCUCUCCUUUCGUAGUCCCCCUCCUCUUAAGUCC-----GCUAA ..(((..(((.((((((((((.....))))))).)))(.(((.((((......................................)))).))).).........))).-----))).. ( -32.36, z-score = -1.92, R) >consensus CCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGAGGGUCUGCUUGGUAUGCCCCUACCCCUGAUAUCCUCCUCCCCUUUCGUAGUCCCACUCCUCUUAAAUCC_____GCUAA ..(((......((((((((((.....))))))).)))..(((.((((......................................)))).)))....................))).. (-20.10 = -20.24 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:56:02 2011