| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,113,801 – 21,113,904 |
| Length | 103 |
| Max. P | 0.927035 |

| Location | 21,113,801 – 21,113,904 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.94 |
| Shannon entropy | 0.70094 |
| G+C content | 0.38170 |
| Mean single sequence MFE | -22.29 |
| Consensus MFE | -9.22 |
| Energy contribution | -8.94 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
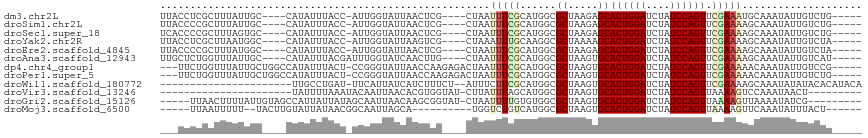
>dm3.chr2L 21113801 103 + 23011544 UUACCUCGCUUUAUUGC----CAUAUUUACC-AUUGGUAUUAACUCG----CUAAUUUCGCAUGGCGCUAAGAGCACUGGAUCUAUCCAGUUCGAAAUGCAAAUAUUGUCUG----- .....(((((((..(((----(((...((((-...)))).......(----(.......))))))))...)))))((((((....))))))..)).................----- ( -22.10, z-score = -1.43, R) >droSim1.chr2L 20693464 103 + 22036055 UUACCCCGCUUUAUUGC----CAUAUUUACC-AUUGGUAUUAACUCG----CUAAUUUCGCAUGGCGCUAAGAGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUUGUCUG----- .......(((((...((----(((...((((-...)))).......(----(.......)))))))((.....))((((((....))))))....)))))............----- ( -22.80, z-score = -1.63, R) >droSec1.super_18 1032142 103 + 1342155 UCACCCCGCUUUAGUGC----CAUAUUUACC-AUUGGUAUUAACUCG----CUAAUUUCGCAUGGCGCUAAGAGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUUGUCUG----- ((.....((((((((((----(((...((((-...)))).......(----(.......)))))))))).)))))((((((....))))))..)).................----- ( -27.80, z-score = -2.97, R) >droYak2.chr2R 19483718 103 - 21139217 UUACCUCGCUUAAUGGC----CAUAUUUACC-AUUGGUAUUAAGUCG----CUAAAUUUGCAAGGCGCUAAAAGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUUGUCUA----- ......(((((((((.(----((........-..)))))))))).))----....((((((.....((.....))((((((....)))))).......))))))........----- ( -22.90, z-score = -1.28, R) >droEre2.scaffold_4845 19209875 103 - 22589142 UUACCCCGCUUUAUGGC----CAUAUUUACC-AUUGGUAUUAACUCG----CUAAUUUCGCAUGGCGCUAAGAGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUUGUCUA----- .......(((((.((((----(((...((((-...)))).......(----(.......)))))))((.....))((((((....)))))).)).)))))............----- ( -23.40, z-score = -1.28, R) >droAna3.scaffold_12943 1413434 104 - 5039921 UUGCUCUGGUUUAUUGC----CAUAUUUACGAUUUGGUAUCAACUUG----CUAAUUUCGCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUUGUCAU----- (((((....((((.(((----(((.....(((.((((((......))----))))..))).)))))).))))((.((((((....)))))).))...)))))..........----- ( -25.00, z-score = -1.28, R) >dp4.chr4_group1 306219 108 - 5278887 ---UUCUGGUUUAUUGCUGGCCAUAUUUACU-CCGGGUAUUAACCAAGAGACUAAUUUCGCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUCGAAAAACAAAUAUUGUCCG----- ---....(((.....)))(((.((((((.((-(..(((....)))..))).....(((((....((((...))))((((((....)))))).)))))...)))))).)))..----- ( -24.50, z-score = -0.77, R) >droPer1.super_5 244765 108 + 6813705 ---UUCUGGUUUAUUGCUGGCCAUAUUUACU-CCGGGUAUUAACCAAGAGACUAAUUUCGCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUCGAAAAACAAAUAUUGUCUG----- ---....(((.....)))(((.((((((.((-(..(((....)))..))).....(((((....((((...))))((((((....)))))).)))))...)))))).)))..----- ( -24.90, z-score = -1.00, R) >droWil1.scaffold_180772 4026347 92 - 8906247 ----------------------UUGCCUGAU-UUCAUUAUCAUCUUUCU--AUUUCUUCGCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUAUACACAUACA ----------------------((((.((((-......)))).......--.((((...((((........))))((((((....))))))..)))).))))............... ( -16.80, z-score = -1.35, R) >droVir3.scaffold_13246 473524 85 - 2672878 ----------------------UAUUUUAAAUACAAUUAACACGUGGUAU-CUUAUUUAGCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUAAAAGUCCAAAUAACU--------- ----------------------......................(((...-((((((((((.....)))))))..((((((....))))))...))).))).......--------- ( -16.20, z-score = -1.48, R) >droGri2.scaffold_15126 3758843 102 + 8399593 -----UUAACUUUUAUUGUAGCCAUUAUUAUAGCAAUUAACAAGCGGUAU-CUAAUUUUGUGUGGCGCUAAGUGCACUGGAUCUAUCCAGUUAAAAGUUAAAAUAUCG--------- -----((((((((((.................(((.......((((.(((-.((....)).))).))))...)))((((((....)))))))))))))))).......--------- ( -22.00, z-score = -1.60, R) >droMoj3.scaffold_6500 6030563 94 - 32352404 -----UUAAUUUUU--UACUUGUAUUAUAACGGCAAUUAGCA----------UGGUCUGUCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUAAAAGUUCAAAUAUUUACU------ -----.........--.....(((((..(((.(((.(((((.----------..(((......)))))))).)))((((((....)))))).....)))..))))).....------ ( -19.10, z-score = -1.31, R) >consensus ___CCCCGCUUUAUUGC____CAUAUUUACC_AUUGGUAUUAACUCG____CUAAUUUCGCAUGGCGCUAAGUGCACUGGAUCUAUCCAGUUCGAAAAGCAAAUAUUGUCUG_____ ...........................((((....))))................(((((......((.....))((((((....)))))).))))).................... ( -9.22 = -8.94 + -0.28)
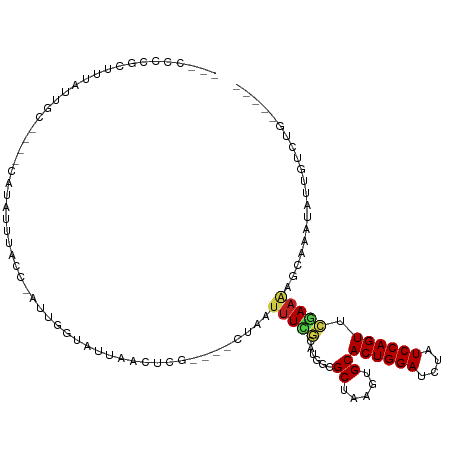
| Location | 21,113,801 – 21,113,904 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.94 |
| Shannon entropy | 0.70094 |
| G+C content | 0.38170 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -10.05 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21113801 103 - 23011544 -----CAGACAAUAUUUGCAUUUCGAACUGGAUAGAUCCAGUGCUCUUAGCGCCAUGCGAAAUUAG----CGAGUUAAUACCAAU-GGUAAAUAUG----GCAAUAAAGCGAGGUAA -----..............(((((..((((((....))))))(((.(((..(((((((.......)----).......((((...-))))...)))----))..))))))))))).. ( -24.30, z-score = -1.13, R) >droSim1.chr2L 20693464 103 - 22036055 -----CAGACAAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCUCUUAGCGCCAUGCGAAAUUAG----CGAGUUAAUACCAAU-GGUAAAUAUG----GCAAUAAAGCGGGGUAA -----.........((((((((....((((((....))))))((.....))(((((((.......)----).......((((...-))))...)))----))...)))))))).... ( -24.40, z-score = -0.72, R) >droSec1.super_18 1032142 103 - 1342155 -----CAGACAAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCUCUUAGCGCCAUGCGAAAUUAG----CGAGUUAAUACCAAU-GGUAAAUAUG----GCACUAAAGCGGGGUGA -----..........(..((((....((((((....))))))(((.((((.(((((((.......)----).......((((...-))))...)))----)).)))))))))))..) ( -27.10, z-score = -1.21, R) >droYak2.chr2R 19483718 103 + 21139217 -----UAGACAAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCUUUUAGCGCCUUGCAAAUUUAG----CGACUUAAUACCAAU-GGUAAAUAUG----GCCAUUAAGCGAGGUAA -----............(((......((((((....))))))......)))(((((((....((((----....))))....(((-(((.......----))))))..))))))).. ( -26.30, z-score = -1.88, R) >droEre2.scaffold_4845 19209875 103 + 22589142 -----UAGACAAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCUCUUAGCGCCAUGCGAAAUUAG----CGAGUUAAUACCAAU-GGUAAAUAUG----GCCAUAAAGCGGGGUAA -----........((((((((((((.((((((....))))))((.....))......)))))..))----)))))...((((.((-(((.......----))))).......)))). ( -24.20, z-score = -0.43, R) >droAna3.scaffold_12943 1413434 104 + 5039921 -----AUGACAAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGCGAAAUUAG----CAAGUUGAUACCAAAUCGUAAAUAUG----GCAAUAAACCAGAGCAA -----..........(((((((....((((((....))))))((.....))(((((((.......)----).....(((.....)))......)))----))........))))))) ( -22.50, z-score = -1.19, R) >dp4.chr4_group1 306219 108 + 5278887 -----CGGACAAUAUUUGUUUUUCGAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGCGAAAUUAGUCUCUUGGUUAAUACCCGG-AGUAAAUAUGGCCAGCAAUAAACCAGAA--- -----.((...(((((((..(((((.((((((....))))))((.....))......))))).....((((.(((....))).))-)))))))))..))...............--- ( -25.30, z-score = -0.94, R) >droPer1.super_5 244765 108 - 6813705 -----CAGACAAUAUUUGUUUUUCGAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGCGAAAUUAGUCUCUUGGUUAAUACCCGG-AGUAAAUAUGGCCAGCAAUAAACCAGAA--- -----.(((((.....)))))((((.((((((....)))))).).....((((((((..........((((.(((....))).))-))....))))))..)).........)))--- ( -22.54, z-score = -0.50, R) >droWil1.scaffold_180772 4026347 92 + 8906247 UGUAUGUGUAUAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGCGAAGAAAU--AGAAAGAUGAUAAUGAA-AUCAGGCAA---------------------- .................(((......((((((....))))))......)))(((......(....)--.......((((......-)))))))..---------------------- ( -17.00, z-score = 0.13, R) >droVir3.scaffold_13246 473524 85 + 2672878 ---------AGUUAUUUGGACUUUUAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGCUAAAUAAG-AUACCACGUGUUAAUUGUAUUUAAAAUA---------------------- ---------...(((.(((..(((((((((((....))))))....(((((.....))))).))))-)..))).)))..................---------------------- ( -15.00, z-score = -0.44, R) >droGri2.scaffold_15126 3758843 102 - 8399593 ---------CGAUAUUUUAACUUUUAACUGGAUAGAUCCAGUGCACUUAGCGCCACACAAAAUUAG-AUACCGCUUGUUAAUUGCUAUAAUAAUGGCUACAAUAAAAGUUAA----- ---------.......((((((((((((((((....))))))((.....))...............-.......((((.....(((((....))))).))))))))))))))----- ( -19.60, z-score = -1.63, R) >droMoj3.scaffold_6500 6030563 94 + 32352404 ------AGUAAAUAUUUGAACUUUUAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGACAGACCA----------UGCUAAUUGCCGUUAUAAUACAAGUA--AAAAAUUAA----- ------......((((((........((((((....))))))(((.(((((...(((......))----------)))))).)))..........))))))--.........----- ( -19.10, z-score = -2.19, R) >consensus _____CAGACAAUAUUUGCUUUUCGAACUGGAUAGAUCCAGUGCACUUAGCGCCAUGCGAAAUUAG____CGAGUUAAUACCAAU_GGUAAAUAUG____GCAAUAAAGCAGGG___ ..............(((((.......((((((....))))))((.....)).....)))))........................................................ (-10.05 = -10.35 + 0.30)

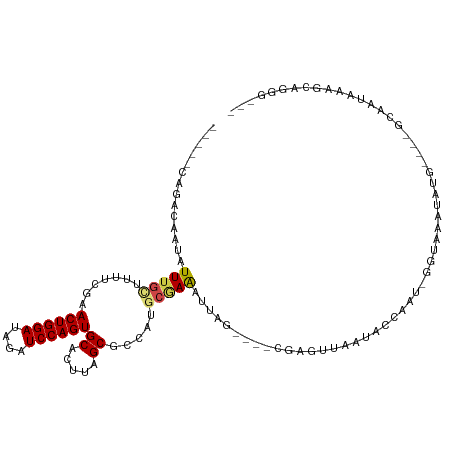
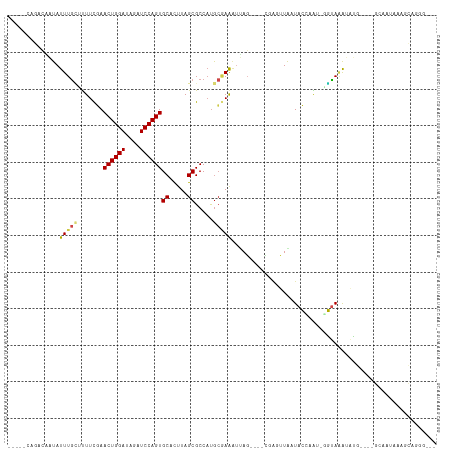
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:56 2011