| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,986,700 – 1,986,796 |
| Length | 96 |
| Max. P | 0.664608 |

| Location | 1,986,700 – 1,986,796 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.52683 |
| G+C content | 0.40905 |
| Mean single sequence MFE | -15.91 |
| Consensus MFE | -7.74 |
| Energy contribution | -7.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1986700 96 + 23011544 --------------GACUUCGAUAAAACAAACCG--CCCACAAGCCAAGGAGCUUCA----UAAAAUGGAAAUCAAAGCACGCGACAGAAGCGUGAUAUGACCACGAAU-ACCCUAC --------------...((((............(--(((.........)).))....----.....(((...(((...(((((.......)))))...)))))))))).-....... ( -17.00, z-score = -2.05, R) >droSec1.super_14 1930493 96 + 2068291 --------------UACUUCGAUAAAACAAACCG--CCCACAAGCCAAGGAGCUCCA----UAAAAUGGAAAUCAGAGCACGCGACAGAAGCGUGAUAUGACCACGAAU-ACCCUAC --------------...((((............(--(((.........)).))((((----(...)))))..(((...(((((.......)))))...)))...)))).-....... ( -19.10, z-score = -2.89, R) >droEre2.scaffold_4929 2032538 96 + 26641161 --------------CACUUCGAUAAAACAAACCG--CCCAUAACCCAAGGAGCUCCA----UAAAAUGGAAAUAAGAGCACGCGACAGAAGCGUGAUAUGGCCACGAAU-ACCCUAC --------------...((((............(--(((.........)).))....----.....(((..(((....(((((.......))))).)))..))))))).-....... ( -17.60, z-score = -1.88, R) >droAna3.scaffold_12916 9702245 98 - 16180835 ------------UGCAGUUCGAUAAAACAAACCG--CCUCCCGAAGAAGUAGUUCCA----UAAAAUAAAUGUCAAAGCACGCGACAAAAGCGUGAUAAGUUUUGGUAU-CCCCUAC ------------..........((((((.(((..--(.((.....)).)..)))...----.................(((((.......)))))....))))))....-....... ( -13.10, z-score = 0.01, R) >dp4.chr4_group3 8758174 105 + 11692001 ------------UGCAGUUCGAUAAAACAAACUUUUCUCAUAAACCAAGAAGUACGAGUAGUACCAUAAAAAUCAAAGCACGCGACCAAAGCGUGAUAUGUCAAUGUAUUACCCUAC ------------((((....((((...........................((((.....))))..............(((((.......)))))...))))..))))......... ( -14.90, z-score = -0.83, R) >droPer1.super_1 10211071 105 + 10282868 ------------UGCAGUUCGAUAAAACAAACUUUUCUCAUAAACCAAGAAGUACGAGUAGUACCAUAAAAAUCAAAGCACGCGACCAAAGCGUGAUAUGUCAAUGUAUUACCCUAC ------------((((....((((...........................((((.....))))..............(((((.......)))))...))))..))))......... ( -14.90, z-score = -0.83, R) >droWil1.scaffold_180772 1220820 111 - 8906247 AUUUCCGAACCAUGCAAGUCGAUAAAAAAAAUAAUGAAGAGUUUAUAAAGAAAAAUA----GUUCAUAAAAAUCAAAACACGCGACAAAAGCGUGAUAUGUCAACG--UUACACUAC .................((.((((.........(((((..((((........)))).----.)))))...........(((((.......)))))...)))).)).--......... ( -14.80, z-score = -0.95, R) >consensus ____________UGCACUUCGAUAAAACAAACCG__CCCACAAACCAAGGAGCUCCA____UAAAAUAAAAAUCAAAGCACGCGACAAAAGCGUGAUAUGUCAACGAAU_ACCCUAC ..............................................................................(((((.......)))))...................... ( -7.74 = -7.74 + 0.00)

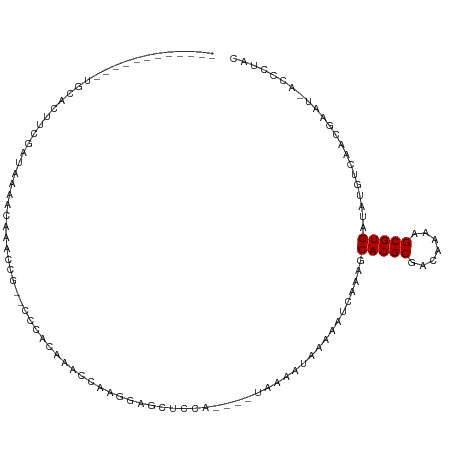

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:46 2011