
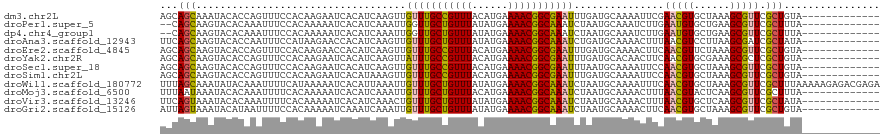
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,104,655 – 21,104,762 |
| Length | 107 |
| Max. P | 0.733818 |

| Location | 21,104,655 – 21,104,762 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Shannon entropy | 0.30570 |
| G+C content | 0.36272 |
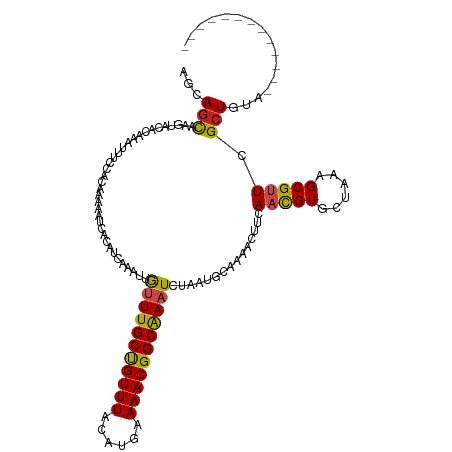
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -16.54 |
| Energy contribution | -16.07 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.733818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 21104655 107 + 23011544 AGCAGCAAAUACACCAGUUUCCACAAGAAUCACAUCAAGUUGUUUGCCGUUUACAUGAAAACGGCGAAUUUGAUGCAAAAUUCGAACGUGCUAAAGCGUUCGCUGUA------------- .(((((....................((((..((((((...(((((((((((......)))))))))))))))))....))))((((((......))))))))))).------------- ( -36.00, z-score = -4.44, R) >droPer1.super_5 231709 105 + 6813705 --CAGCAAGUACACAAAUUUCCACAAAAAUCACAUCAAAUUGGUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAUCUUGAAUGUGCUGAAGCGUUCGCUUUA------------- --.....(((((((((((((.((.........((......)).(((((((((......)))))))))......)).)))).)))..))))))(((((....))))).------------- ( -20.40, z-score = -0.56, R) >dp4.chr4_group1 293086 105 - 5278887 --CAGCAAGUACACAAAUUUCCACAAAAAUCACAUCAAAUUGGUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAUCUUGAAUGUGCUGAAGCGUUCGCUUUA------------- --.....(((((((((((((.((.........((......)).(((((((((......)))))))))......)).)))).)))..))))))(((((....))))).------------- ( -20.40, z-score = -0.56, R) >droAna3.scaffold_12943 1405528 107 - 5039921 UUCAGCAAGUACACCAAUUUCCAUAAGAACCACAUCAAGUUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUGAUGCAAAACUUUAACGUCCUUAAGCGAUCGCUAUA------------- ...(((..........................(((((....(((((((((((......))))))))))).)))))...........(((......)))...)))...------------- ( -19.90, z-score = -1.38, R) >droEre2.scaffold_4845 19201654 107 - 22589142 AGCAGCAAGUACACCAGUUUCCACAAGAACCACAUCAAGUUGUUUGCCGUUUACAUGAAAACGGCGAAUUUGAUGCAAAACUUCAACGUUCUAAAGCGUUCGCUGUA------------- .(((((((((......((((......))))..((((((...(((((((((((......)))))))))))))))))....)))).(((((......))))).))))).------------- ( -30.80, z-score = -3.11, R) >droYak2.chr2R 19476299 107 - 21139217 AGCAGCAAGUACACCAGUUUCCACAAGAAUCACAUCAAGUUAUUUGCCGUUUACAUGAAAACGGCGAAUUUGAUGCACAACUUCAACGUGCGAAAGCGCUCGCUGUA------------- .(((((((((......(.(((.....))).).((((((((...(((((((((......)))))))))))))))))....))))....((((....))))..))))).------------- ( -35.20, z-score = -4.02, R) >droSec1.super_18 1023639 107 + 1342155 AGCAGCAAGUACACCAGUUUCCACAAGAAUCACAUCAAGUUGUUUGCCGUUUACAUGAAAACGGCGAAUUUAAUGCAAAAUUCCAACGUGCUAAAGCGUUCGCUGUA------------- .(((((.(((((....(.(((.....))).).......((((...(((((((......)))))))((((((......)))))))))))))))...(....)))))).------------- ( -25.80, z-score = -1.68, R) >droSim1.chr2L 20682449 107 + 22036055 AGCAGCAAGUACACCAGUUUCCACAAGAAUCACAUAAAGUUGUUUGCCGUUUACAUGAAAACGGCGAAUUUGAUGCAAAAUUCCAACGUGCUAAAGCGUUCGCUGUA------------- .(((((.(((((....(.(((.....))).).......((((...(((((((......)))))))((((((......)))))))))))))))...(....)))))).------------- ( -26.70, z-score = -1.56, R) >droWil1.scaffold_180772 4008276 120 - 8906247 UUUAGCAAAUAUACAAAUUUUCAUAAAAAUCACAUUAAAUUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAAUUUCAACGUGCUAAAGCGUUCGCUUUAAAAAGAGACGAGA (((((((.......((((((((((.......(((......)))(((((((((......))))))))).....))).))))))).....)))))))...((((((((....)))).)))). ( -24.10, z-score = -1.66, R) >droMoj3.scaffold_6500 6019539 107 - 32352404 UUUAAUAAAUACACAAAUUUUCACAAAAAUCACAUCAAAUUGUUUGCUGUUUACAUGAAAACGGCAAAUCUAAUGCAAAACUUUAACGUACUCAAGCGUUCGCUUUA------------- .........................................(((((((((((......)))))))))))...............(((((......))))).......------------- ( -14.00, z-score = -0.85, R) >droVir3.scaffold_13246 452365 107 - 2672878 UUCAGUAAAUACACAAAUUUUCACAAAAAUCACAUCAAACUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAACUUUAACGUGCUCAAGCGUUCGCUAUA------------- ...(((...................................(((((((((((......)))))))))))...............(((((......))))).)))...------------- ( -15.30, z-score = -0.62, R) >droGri2.scaffold_15126 3747810 107 + 8399593 AUUAGUAAAUACAUAAUUUUCCACAAAAAUCAAAUCAAAUUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAACUUCAACGUGCUAAAGCGUUCGCUGUA------------- ..((((...................................(((((((((((......)))))))))))...............(((((......))))).))))..------------- ( -16.20, z-score = -0.38, R) >consensus AGCAGCAAGUACACAAAUUUCCACAAAAAUCACAUCAAAUUGUUUGCUGUUUACAUGAAAACGGCAAAUCUAAUGCAAAACUUCAACGUGCUAAAGCGUUCGCUGUA_____________ ...(((...................................(((((((((((......)))))))))))...............(((((......))))).)))................ (-16.54 = -16.07 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:53 2011