| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,101,820 – 21,101,934 |
| Length | 114 |
| Max. P | 0.696749 |

| Location | 21,101,820 – 21,101,934 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.83 |
| Shannon entropy | 0.67006 |
| G+C content | 0.51713 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -10.23 |
| Energy contribution | -9.71 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
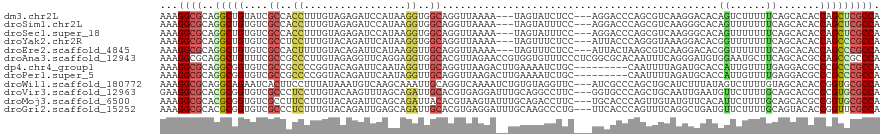
>dm3.chr2L 21101820 114 + 23011544 AAAGGCGCAGGCUGUAUCGCCACCUUUGUAGAGAUCCAUAAGGUGGCAGGUUAAAA---UAGUAUCUCC---AGGACCCAGCGUCAAGGACACAGUCUUUUUCAGCACACCAGCUCGCCA ...((((.(((((((...((((((((((........)).)))))))).........---.......(((---..(((.....)))..))).))))))).....(((......))))))). ( -36.00, z-score = -2.56, R) >droSim1.chr2L 20679676 114 + 22036055 AAAGGCGCAGGCUGUGUCGCCACCUUUGUAGAGAUCCAUAAGGUGGCAGGUUAAAA---UAGUAUUUCC---AGGACCCAGCGUCAAGGGCACAGUUUUUUUCAGCACACCAGCUCGCCA ...((((..(((((((((((((((((((........)).)))))))).((..((((---(.((...(((---..(((.....)))..))).)).)))))..)).).))).))))))))). ( -38.00, z-score = -2.58, R) >droSec1.super_18 1020821 114 + 1342155 AAAGGCGCAGGCUGUGUCGCCACCUUUGUAGAGAUCCAUAAGGUGGCAGGUUAAAA---UAGUAUUUCC---AGGACCCAGCGUCAAGGGCACAGUUUUUUUCAGCACACCAGCUCGCCA ...((((..(((((((((((((((((((........)).)))))))).((..((((---(.((...(((---..(((.....)))..))).)).)))))..)).).))).))))))))). ( -38.00, z-score = -2.58, R) >droYak2.chr2R 19473366 114 - 21139217 AAAGGCGCAGGCUGUGUCGCCUCCUUUGUACAGAUUCAUAAGGUGGCAGGUUAAAA---UAGUUUCUCC---AUUACCCAGGGUAAAGGACACGGUUUUUUUCAGCACACCAGCCCGCCA ...((((..((((((((((((.((((((........)).)))).))).((..((((---(.((...(((---.(((((...))))).))).)).)))))..)).).))).))))))))). ( -38.40, z-score = -2.78, R) >droEre2.scaffold_4845 19198732 114 - 22589142 AAAGGCGCAGGCUGUGUCGCCACUUUUGUACAGAUUCAUAAGGUUGCAGGUUAAAA---UAGUUUCUCC---AUUACUAAGCGUCAAGGACACGGUUUUUUUCAGCACACCAGCCCGCCA ...((((..(((((((((((.(((((((........)).))))).)).((..((((---(.((...(((---...((.....))...))).)).)))))..)).).))).))))))))). ( -27.60, z-score = -0.58, R) >droAna3.scaffold_12943 1402494 120 - 5039921 AAAGGCGCAGGCUGUUUCGCCGCCCUUGUAGAGGUUCAGGAGGUGGCACGUUAGAACCGUGGUGUUUCCCUCGGCGCACAAUUUCAGGGAUGUGGAAUGCUUCAGCACGCCAGCCGCCCA ...((.((.((((((..(((((((((((........)))).))))))......(..(((.((......)).)))..).........)..))..((..(((....)))..)))))))))). ( -43.10, z-score = 0.13, R) >dp4.chr4_group1 289043 111 - 5278887 AAAGGCGCAGGCGGUGUCGCCGCCCCGGUACAGAUUCAAUAGGUUGCAGGUUAAGACUUGAAAAUCUGC---------CAAUUUUAGAUGCACCAUUGUUUUGAGGACGCCCGCCCGCCA ...((((..(((((((((((((...)))).((((..((((.(((.(((..((((((.(((.........---------))))))))).)))))))))).))))..)))).))))))))). ( -38.90, z-score = -1.84, R) >droPer1.super_5 227780 111 + 6813705 AAAGGCGCAGGCGGUGUCGCCGCCCCGGUACAGAUUCAAUAGGUUGCAGGUUAAGACUUGAAAAUCUGC---------CAAUUUUAGAUGCACCAUUGUUUUGAGGACGCCCGCCCGCCA ...((((..(((((((((((((...)))).((((..((((.(((.(((..((((((.(((.........---------))))))))).)))))))))).))))..)))).))))))))). ( -38.90, z-score = -1.84, R) >droWil1.scaffold_180772 4003112 117 - 8906247 AAAGGCGCAGGCAGAAUCACUUCCUUUAUAAAUGUCAAGCAAAUUGCAGGUCAAAAUCUGUGUAGGUUC---AUCGCCCAGCUGCAUCUUUAUAGUCUUUUGUAGCACACCGGUGCGCCA ...(((((((((.(((((..............((.....))....((((((....))))))...)))))---...)))..((((((..............)))))).......)))))). ( -32.04, z-score = -1.56, R) >droVir3.scaffold_12963 9685241 117 - 20206255 GAAGGCGCACGCGGUGUCGCCCUCCUUGUACAAGUUUAGCAGAUUGCACGUGAGGAUUUGCAGGCCUUC---GGUGCCCAGCUGCAAUUGAAUGUUCUUUUGCAGCACGCCCGUGCGCCA ...((((((((.(((((.((..((((..(...((((.....))))....)..))))...)).((((...---.).)))..(((((((..((....))..)))))))))))))))))))). ( -52.20, z-score = -3.41, R) >droMoj3.scaffold_6500 13120888 117 - 32352404 AAAGGCGCACGCGGUGUCGCCUUCCUUGUACAGAUUCAGCAGAUUACACGUAAGUAUUUGCAGACCUUC---UGCACCCAGUUGUAUGUUCACAUUCUUUUGCAGCACGCCGGUGCGCCA ...(((((((.((((((..........((.((((.((.((((((...((....)))))))).))...))---)).))...((((((..............))))))))))))))))))). ( -41.64, z-score = -3.26, R) >droGri2.scaffold_15252 8620052 117 - 17193109 AAAGGCGCACGCGGUGUCGCCCUCUUUGUACAGAUUGAGCAGAUUGCACGUGAGGAUUUGCAAGCCCUG---UUCACCCAGUUUCAGGCUGAUGUUCUUUUGCAGUACACCGGUUCGCCA ...((((.((.((((((((((.(((......))).(((((((.((((((....)....)))))...)))---))))..........)))...(((......)))).)))))))).)))). ( -38.00, z-score = -0.92, R) >consensus AAAGGCGCAGGCUGUGUCGCCACCUUUGUACAGAUUCAGAAGGUUGCAGGUUAAAA__UGAGUAUCUCC___AGCACCCAGCUUCAAGGGCACAGUUUUUUUCAGCACACCAGCCCGCCA ...((((..(((((....((..((.................))..))..............................................((......)).......))))))))). (-10.23 = -9.71 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:52 2011