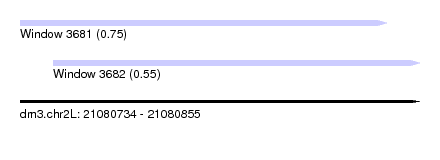
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 21,080,734 – 21,080,855 |
| Length | 121 |
| Max. P | 0.748672 |

| Location | 21,080,734 – 21,080,845 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Shannon entropy | 0.39299 |
| G+C content | 0.55297 |
| Mean single sequence MFE | -41.19 |
| Consensus MFE | -19.79 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
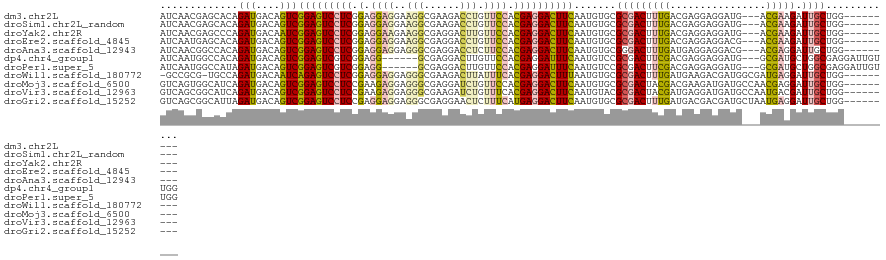
>dm3.chr2L 21080734 111 + 23011544 AUCAACGAGCACAGAUGACAGUCGGAGUCCUCGGAGGAGGAAGGCGAAGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG---ACGAAGAUUGCUGG--------- ........((((((((....)))((((((((((..((((..(((......))).)))).))))))))))..)))))(((((((((.((.......))---.))))).))))...--------- ( -40.80, z-score = -2.65, R) >droSim1.chr2L_random 866571 111 + 909653 AUCAACGAGCACAGAUGACAGUCGGAGUCCUCGGAGGAGGAAGGCGAAGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG---ACGAAGAUUGCUGG--------- ........((((((((....)))((((((((((..((((..(((......))).)))).))))))))))..)))))(((((((((.((.......))---.))))).))))...--------- ( -40.80, z-score = -2.65, R) >droYak2.chr2R 19452895 111 - 21139217 AUCAACGAGCCCAGAUGACAAUCGGAGUCCUCGGAGGAAGAAGGCGAGGACUUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG---ACGAAGAUUGCUGG--------- ..........((((....(((((((((((((((..((((.(((.......))).)))).))))))))))......((((.((((......)))).))---.))..)))))))))--------- ( -37.90, z-score = -1.95, R) >droEre2.scaffold_4845 19177893 111 - 22589142 AUCAAUGAGCACAGAUGACAGUCGGAGUCCUCGGAGGAGGAAGGCGAGGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGACG---ACGAAGAUUGCUGG--------- ........((((((((....)))((((((((((..((((..(((......))).)))).))))))))))..)))))(((((((((.((.......))---.))))).))))...--------- ( -43.10, z-score = -3.16, R) >droAna3.scaffold_12943 1379678 111 - 5039921 AUCAACGGCCACAGAUGACAGUCGGAGUCCUCGGAGGAGGAGGGCGAGGACCUCUUCCACGAGGACUUCAAUGUGCGGGACUUUGAUGAGGAGGACG---ACGAGGAUUGCUGG--------- .....(((((((((((....)))((((((((((..((((((((.......)))))))).))))))))))..))))..(..((((.....))))..).---.........)))).--------- ( -45.90, z-score = -2.91, R) >dp4.chr4_group1 264280 114 - 5278887 AUCAAUGGCCACAGAUGACAGUCGGAGUCGUCGGAGG------GCGAGGACUUGUUCCACGAGGAUUUCAAUGUCCGCGACUUCGACGAGGAGGAUG---GCGAUGCUGGCGAGGAUUGUUGG ....((.((((.........((((((((((.((((.(------..((((.((((.....))))..))))..).))))))))))))))........))---)).)).(..((((...))))..) ( -39.63, z-score = -0.70, R) >droPer1.super_5 202978 114 + 6813705 AUCAAUGGCCAUAGAUGACAGUCGGAGUCGUCGGAGG------GCGAGGACUUGUUCCACGAGGAUUUCAAUGUCCGCGACUUCGACGAGGAGGAUG---GCGAUGCUGGCGAGGAUUGUUGG ....((.(((((........((((((((((.((((.(------..((((.((((.....))))..))))..).)))))))))))))).......)))---)).)).(..((((...))))..) ( -41.96, z-score = -1.60, R) >droWil1.scaffold_180772 3970482 112 - 8906247 -GCCGCG-UGCCAGAUGACAAUCAGAGUCCUCGGAGGAGGAGGGCGAAGACUUAUUUCACGAGGACUUUAAUGUGCGCGACUUUGAUGAAGACGAUGGCGAUGAGGAUUGCUGG--------- -..((((-..(..(((....)))((((((((((..(((.(((........))).)))..))))))))))...)..)))).................((((((....))))))..--------- ( -36.60, z-score = -1.82, R) >droMoj3.scaffold_6500 13094996 114 - 32352404 GUCAGUGGCAUCAGAUGACAGUCGGAGUCCUCCGAAGAGGAGGGCGAGGAUCUGUUCCACGAGGACUUCAAUGUGCGCGACUACGACGAAGAUGAUGCCAACGAGGAUUGCUGG--------- .((..((((((((..(....((((..(((((((.....)))))))..(((.....))).((.(.((......)).).))....))))..)..))))))))..))..........--------- ( -40.90, z-score = -1.36, R) >droVir3.scaffold_12963 9660962 114 - 20206255 GUCAGCGGCAUCAGAUGACAGUCGGAGUCCUCCGAAGAGGAGGGCGAAGAUCUGUUUCACGAGGACUUCAAUGUACGCGACUACGAUGAGGAUGAUGCCAAUGACGAUUGCUGG--------- ((((..(((((((((.(((((((...(((((((.....)))))))...)).))))))).......(((((.((((......)))).))))).)))))))..)))).........--------- ( -46.00, z-score = -3.51, R) >droGri2.scaffold_15252 8597338 114 - 17193109 GUCAGCGGCAUUAGAUGACAGUCGGAGUCCUCCGAGGAGGAGGGCGAGGAACUCUUUCAUGAGGACUUCAAUGUGCGCGACUUUGAUGACGACGAUGCUAAUGAGGAUUGCUGG--------- ..((((((((((((......((((..(((((((.....)))))))((((..(((......)))..))))....................))))....))))))....)))))).--------- ( -39.50, z-score = -1.60, R) >consensus AUCAACGGCCACAGAUGACAGUCGGAGUCCUCGGAGGAGGAGGGCGAGGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG___ACGAAGAUUGCUGG_________ .............(((....)))(((((((((.(.((((..(((......))).)))).)))))))))).......(((((((((................))))).))))............ (-19.79 = -19.92 + 0.13)

| Location | 21,080,744 – 21,080,855 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.47 |
| Shannon entropy | 0.31685 |
| G+C content | 0.54048 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
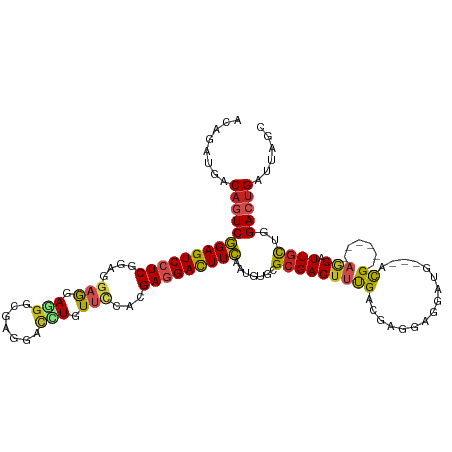
>dm3.chr2L 21080744 111 + 23011544 ACAGAUGACAGUCGGAGUCCUCGGAGGAGGAAGGCGAAGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG---ACG------AAGAUUGCUGGACUGAUUAGC ........(((((((((((((((..((((..(((......))).)))).)))))))))).......(((((((((.((.......))---.))------))).))))..)))))...... ( -39.70, z-score = -2.79, R) >droSim1.chr2L_random 866581 111 + 909653 ACAGAUGACAGUCGGAGUCCUCGGAGGAGGAAGGCGAAGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG---ACG------AAGAUUGCUGGACUGACUAGA ........(((((((((((((((..((((..(((......))).)))).)))))))))).......(((((((((.((.......))---.))------))).))))..)))))...... ( -39.70, z-score = -2.59, R) >droYak2.chr2R 19452905 111 - 21139217 CCAGAUGACAAUCGGAGUCCUCGGAGGAAGAAGGCGAGGACUUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG---ACG------AAGAUUGCUGGACUGACUAGC ((((....(((((((((((((((..((((.(((.......))).)))).))))))))))......((((.((((......)))).))---.))------..))))))))).......... ( -38.70, z-score = -2.71, R) >droEre2.scaffold_4845 19177903 111 - 22589142 ACAGAUGACAGUCGGAGUCCUCGGAGGAGGAAGGCGAGGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGACG---ACG------AAGAUUGCUGGACUGACUAGC ........(((((((((((((((..((((..(((......))).)))).)))))))))).......(((((((((.((.......))---.))------))).))))..)))))...... ( -42.00, z-score = -2.83, R) >droAna3.scaffold_12943 1379688 111 - 5039921 ACAGAUGACAGUCGGAGUCCUCGGAGGAGGAGGGCGAGGACCUCUUCCACGAGGACUUCAAUGUGCGGGACUUUGAUGAGGAGGACG---ACG------AGGAUUGCUGGACUGAUUAGC ........(((((((((((((((..((((((((.......)))))))).)))))))))).....((((..(((((.((.......))---.))------))).))))..)))))...... ( -46.70, z-score = -4.02, R) >dp4.chr4_group1 264290 114 - 5278887 ACAGAUGACAGUCGGAGUCGUC------GGAGGGCGAGGACUUGUUCCACGAGGAUUUCAAUGUCCGCGACUUCGACGAGGAGGAUGGCGAUGCUGGCGAGGAUUGUUGGACUGAUUAGC ........(((((....(((((------((((..((.(((((((.(((....)))...))).)))).)).))))))))).......((((((.((....)).)))))).)))))...... ( -41.00, z-score = -1.69, R) >droPer1.super_5 202988 114 + 6813705 AUAGAUGACAGUCGGAGUCGUC------GGAGGGCGAGGACUUGUUCCACGAGGAUUUCAAUGUCCGCGACUUCGACGAGGAGGAUGGCGAUGCUGGCGAGGAUUGUUGGACUGAUUAGC ........(((((....(((((------((((..((.(((((((.(((....)))...))).)))).)).))))))))).......((((((.((....)).)))))).)))))...... ( -41.00, z-score = -1.89, R) >droWil1.scaffold_180772 3970491 113 - 8906247 -CAGAUGACAAUCAGAGUCCUCGGAGGAGGAGGGCGAAGACUUAUUUCACGAGGACUUUAAUGUGCGCGACUUUGAUGAAGACGAUGGCGAUG------AGGAUUGCUGGACUGAUUAGC -(((.(..(((((((((((((((..(((.(((........))).)))..))))))))))....(((.((.((((...)))).))...)))...------..)))))..)..)))...... ( -30.10, z-score = -1.35, R) >droMoj3.scaffold_6500 13095006 114 - 32352404 UCAGAUGACAGUCGGAGUCCUCCGAAGAGGAGGGCGAGGAUCUGUUCCACGAGGACUUCAAUGUGCGCGACUACGACGAAGAUGAUGCCAACG------AGGAUUGCUGGACCGAUUAGC .........((((((.(((((((.....)))))))..........(((((((...((((..((.(((((.((.......)).)).)))))..)------))).))).))))))))))... ( -32.80, z-score = 0.22, R) >droVir3.scaffold_12963 9660972 114 - 20206255 UCAGAUGACAGUCGGAGUCCUCCGAAGAGGAGGGCGAAGAUCUGUUUCACGAGGACUUCAAUGUACGCGACUACGAUGAGGAUGAUGCCAAUG------ACGAUUGCUGGACUGACUAGC (((((.(((((((...(((((((.....)))))))...)).))))))).......(((((.((((......)))).))))).)))........------......(((((.....))))) ( -33.00, z-score = -0.58, R) >droGri2.scaffold_15252 8597348 114 - 17193109 UUAGAUGACAGUCGGAGUCCUCCGAGGAGGAGGGCGAGGAACUCUUUCAUGAGGACUUCAAUGUGCGCGACUUUGAUGACGACGAUGCUAAUG------AGGAUUGCUGGACUGAUUAGC ....((.(.(((((..(((((((.....)))))))((((..(((......)))..))))........))))).).)).........((((((.------((..(....)..)).)))))) ( -36.30, z-score = -1.77, R) >consensus ACAGAUGACAGUCGGAGUCCUCGGAGGAGGAGGGCGAGGACCUGUUCCACGAGGACUUCAAUGUGCGCGACUUUGACGAGGAGGAUG___ACG______AGGAUUGCUGGACUGAUUAGC ........((((((((((((((....(((.(((.......))).)))...))))))))).......(((((((..........................))).))))..)))))...... (-21.30 = -21.63 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:51 2011