
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,986,505 – 1,986,601 |
| Length | 96 |
| Max. P | 0.998742 |

| Location | 1,986,505 – 1,986,601 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 59.43 |
| Shannon entropy | 0.79764 |
| G+C content | 0.42851 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -11.73 |
| Energy contribution | -11.92 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.998742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
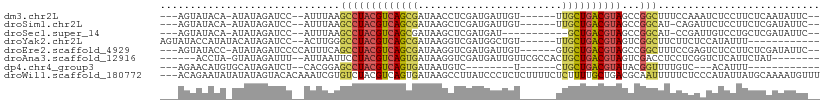
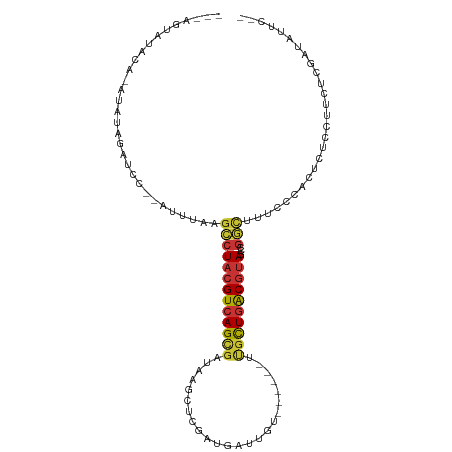
>dm3.chr2L 1986505 96 + 23011544 ---AGUAUACA-AUAUAGAUCC--AUUUAAGCCUACGUCAGCGAUAACCUCGAUGAUUGU------UUGCUGACGUAGCCGGCUUUCCAAAUCUCCUUCUCAAUAUUC-- ---........-....((((..--....(((((((((((((((((((.(.....).))).------)))))))))))...))))).....))))..............-- ( -23.20, z-score = -3.17, R) >droSim1.chr2L 1956161 95 + 22036055 ---AGUAUACA-AUAUAGAUCC--AUUUAAGCCUACGUCAGCGAUAAGCUCGAUGAUUGU------UUGCUGACGUAGCCGGCAU-CAGAUUCUCCUUCUCGAUAUUC-- ---........-..........--......(((((((((((((((((.(.....).))).------)))))))))))...)))((-((((.......))).)))....-- ( -22.50, z-score = -1.26, R) >droSec1.super_14 1930299 90 + 2068291 ---AGUAUACA-AUAUAGAUCC--AUUUAAGCCUACGUCAGCGAUAAGCUCGAUGAU-----------GCUGACGUAGCCGGCAU-CCGAUUGUCCUGCUCGAUAUUC-- ---((((.(((-((...(((((--......(.(((((((((((.((.......)).)-----------)))))))))).))).))-)..)))))..))))........-- ( -21.80, z-score = -0.77, R) >droYak2.chr2L 1968105 90 + 22324452 AGUAUACCAUAUACAUAGAUCC--ACUUGGGCCUACGUCAGCGAUAAGGUCGAUGGCUGU------UUGCUGACGUAGUCGGCUUCUUCUCCAUAUUU------------ .(((((...)))))........--....((((((((((((((((...(((.....)))..------)))))))))))...))))).............------------ ( -26.60, z-score = -1.89, R) >droEre2.scaffold_4929 2032331 98 + 26641161 ---AGUAUACC-AUAUAGAUCCCCAUUUCAGCCUACGUCAGCGAUAAGGUCGAUGAUUGU------GUGCUGACGUAGCCGGCUUUCCGAGUCUCCUUCUCGAUAUUC-- ---......((-.....((........)).(.((((((((((.(((..(((...)))..)------)))))))))))).))).....((((.......))))......-- ( -25.30, z-score = -1.35, R) >droAna3.scaffold_12916 9702033 93 - 16180835 ------ACCUA-GUAUAGAUUU--AUUAAUUCCUACGUCAGUGAUAAGGUCGAUGAUUGUUCGCCACUGCUGACGUAGUCGACCUCCUCGGUCUCAUUCUAU-------- ------.....-..(((((...--........(((((((((((....((.(((.......)))))..)))))))))))..((((.....))))....)))))-------- ( -26.60, z-score = -2.72, R) >dp4.chr4_group3 8757989 76 + 11692001 ---AGAACAUGUGCAUAGAUCU--CACGGAGCCUACGUCAGUGAUAAUGUC--------U------CUGCUGACGUAUACGGUUUUGUC---ACAUUU------------ ---.....(((((.........--.(((((((((((((((((((.......--------)------).)))))))))...)))))))))---))))..------------ ( -21.70, z-score = -1.54, R) >droWil1.scaffold_180772 1220577 107 - 8906247 ---ACAGAAUAUAUAUAGUACACAAAUCGUGUCUACGUCAGUGAUAAGCCUUAUCCCUCUCUUUUCUCUUUUGCUGACGCAAUUUUUCUCCCAUAUUAUGCAAAAUGUUU ---.((.(((((...(((.((((.....)))))))((((((..(..........................)..)))))).............))))).)).......... ( -15.87, z-score = -1.60, R) >consensus ___AGUAUACA_AUAUAGAUCC__AUUUAAGCCUACGUCAGCGAUAAGCUCGAUGAUUGU______UUGCUGACGUAGCCGGCUUUCCCACUCUCCUUCUCGAUAUUC__ ..............................(((((((((((((........................))))))))))...)))........................... (-11.73 = -11.92 + 0.19)
| Location | 1,986,505 – 1,986,601 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 59.43 |
| Shannon entropy | 0.79764 |
| G+C content | 0.42851 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -9.41 |
| Energy contribution | -10.41 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
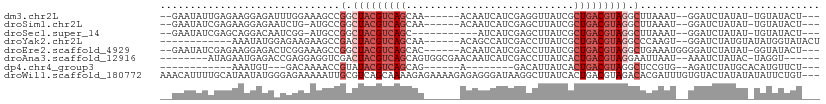
>dm3.chr2L 1986505 96 - 23011544 --GAAUAUUGAGAAGGAGAUUUGGAAAGCCGGCUACGUCAGCAA------ACAAUCAUCGAGGUUAUCGCUGACGUAGGCUUAAAU--GGAUCUAUAU-UGUAUACU--- --..............((((..(..(((((...(((((((((..------..((((.....))))...))))))))))))))...)--..))))....-........--- ( -25.60, z-score = -2.09, R) >droSim1.chr2L 1956161 95 - 22036055 --GAAUAUCGAGAAGGAGAAUCUG-AUGCCGGCUACGUCAGCAA------ACAAUCAUCGAGCUUAUCGCUGACGUAGGCUUAAAU--GGAUCUAUAU-UGUAUACU--- --..(((.(((.....(((.(((.-.....(.((((((((((..------..................)))))))))).)......--))))))...)-)))))...--- ( -20.95, z-score = -0.15, R) >droSec1.super_14 1930299 90 - 2068291 --GAAUAUCGAGCAGGACAAUCGG-AUGCCGGCUACGUCAGC-----------AUCAUCGAGCUUAUCGCUGACGUAGGCUUAAAU--GGAUCUAUAU-UGUAUACU--- --...........((.(((((.((-((.(((.((((((((((-----------...............)))))))))).)......--))))))..))-)))...))--- ( -23.26, z-score = -0.53, R) >droYak2.chr2L 1968105 90 - 22324452 ------------AAAUAUGGAGAAGAAGCCGACUACGUCAGCAA------ACAGCCAUCGACCUUAUCGCUGACGUAGGCCCAAGU--GGAUCUAUGUAUAUGGUAUACU ------------..(((((((......(((...(((((((((..------..................))))))))))))((....--)).)))))))............ ( -22.75, z-score = -1.19, R) >droEre2.scaffold_4929 2032331 98 - 26641161 --GAAUAUCGAGAAGGAGACUCGGAAAGCCGGCUACGUCAGCAC------ACAAUCAUCGACCUUAUCGCUGACGUAGGCUGAAAUGGGGAUCUAUAU-GGUAUACU--- --..((((((....(((..((((...((((...(((((((((..------..................)))))))))))))....))))..)))...)-)))))...--- ( -29.25, z-score = -2.03, R) >droAna3.scaffold_12916 9702033 93 + 16180835 --------AUAGAAUGAGACCGAGGAGGUCGACUACGUCAGCAGUGGCGAACAAUCAUCGACCUUAUCACUGACGUAGGAAUUAAU--AAAUCUAUAC-UAGGU------ --------(((((....((((.....))))..(((((((((....(((((.......))).))......)))))))))........--...)))))..-.....------ ( -27.40, z-score = -2.75, R) >dp4.chr4_group3 8757989 76 - 11692001 ------------AAAUGU---GACAAAACCGUAUACGUCAGCAG------A--------GACAUUAUCACUGACGUAGGCUCCGUG--AGAUCUAUGCACAUGUUCU--- ------------..((((---(.((.....((.((((((((..(------(--------.......)).)))))))).))..(...--.).....))))))).....--- ( -19.20, z-score = -1.35, R) >droWil1.scaffold_180772 1220577 107 + 8906247 AAACAUUUUGCAUAAUAUGGGAGAAAAAUUGCGUCAGCAAAAGAGAAAAGAGAGGGAUAAGGCUUAUCACUGACGUAGACACGAUUUGUGUACUAUAUAUAUUCUGU--- .........(((.((((((..((.....((((((((((......)..........((((.....)))).)))))))))((((.....)))).))...)))))).)))--- ( -19.00, z-score = -0.57, R) >consensus __GAAUAUCGAGAAGGAGAAUCGGAAAGCCGGCUACGUCAGCAA______ACAAUCAUCGACCUUAUCGCUGACGUAGGCUUAAAU__GGAUCUAUAU_UAUAUACU___ ..............................(.(((((((((............................))))))))).).............................. ( -9.41 = -10.41 + 1.00)
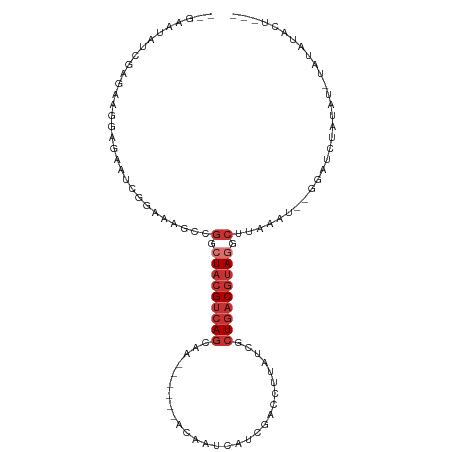
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:46 2011