| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,953,569 – 20,953,645 |
| Length | 76 |
| Max. P | 0.747095 |

| Location | 20,953,569 – 20,953,645 |
|---|---|
| Length | 76 |
| Sequences | 9 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 68.37 |
| Shannon entropy | 0.64522 |
| G+C content | 0.54202 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.17 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
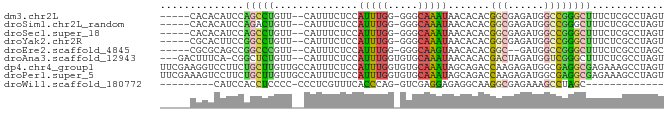
>dm3.chr2L 20953569 76 + 23011544 ACUAGGCGAGAAAGCCCGGCCAUCUCGCCGUGUGUUAUUUGCCC-CCAAAUGGAGAAAUG--AACAGGCUGGAUGUGUG----- ....(((......)))..(((((((.(((.(((.((((((..((-......))..)))))--))))))).))))).)).----- ( -22.80, z-score = -1.25, R) >droSim1.chr2L_random 844034 76 + 909653 ACUAGGCGAGAAAGCCCGGCCAUCUCGCCGUGUGUUAUUUGCCC-CCAAAUGGAGAAAUG--AACAGUCUGGAUGUGUG----- .((((((((((..((...))..))))))).(((.((((((..((-......))..)))))--))))...))).......----- ( -18.80, z-score = -0.47, R) >droSec1.super_18 886654 76 + 1342155 ACUAGGCGAGAAAGCCCGGCCAUCUCGCCGUGUGUUAUUUGCCC-CCAAAUGGAGAAAUG--AACAGGCUGGAUGUGUG----- ....(((......)))..(((((((.(((.(((.((((((..((-......))..)))))--))))))).))))).)).----- ( -22.80, z-score = -1.25, R) >droYak2.chr2R 19339237 76 - 21139217 ACUAGGCGAGAAAGCCCGGCCAUCUCGCCGUGUGUUAUUUGCCC-CCAAAUGGAGAAAUG--AACAGGCCGGAAGUGCG----- ....(((((((..((...))..))))))).(((.((((((..((-......))..)))))--)))).(((....).)).----- ( -22.50, z-score = -0.86, R) >droEre2.scaffold_4845 19071388 74 - 22589142 GCUAGGCGAGAAAGCCCGGCCAUC--GCCGUGUGUUACUUGCCC-CCAAAUGGAGAAAUG--AACGGGCCGGCUGCGCG----- ....(((......)))((((....--)))).((((..((.((((-((....))(....).--...)))).))..)))).----- ( -23.40, z-score = 0.47, R) >droAna3.scaffold_12943 1268576 78 - 5039921 ACUAGGCGAGAAAGCCCGACCAUCUAGUCGUGUGUUAUUUGCACACCAAAUGGAGAAAUG--AACAGAGCCG-UGAAAGUC--- ....(((..........(((......)))((((((.....))))))..............--......))).-........--- ( -15.50, z-score = -0.35, R) >dp4.chr4_group1 88408 84 - 5278887 ACUAGGCUUUCUCGCCUCGCCAUCUCUUGGUCUGCUAUUUGCACACCAAAUGGAGAAAUGGCAACAAGCAGAAGGACCUUCGAA ...(((((((((.((...(((((((((((((.(((.....))).))))...)))))..)))).....)))))))).)))..... ( -28.90, z-score = -3.15, R) >droPer1.super_5 26860 84 + 6813705 ACUAGGCUUUCUCGCCUCGCCAUCUCUUGGUCUGCUAUUUGCACACCAAAUGGAGAAAUGGCAACAAGCAGAAGGACUUUCGAA ...(((((((((.((...(((((((((((((.(((.....))).))))...)))))..)))).....)))))))).)))..... ( -26.20, z-score = -2.35, R) >droWil1.scaffold_180772 3823017 60 - 8906247 -------------GCUAGGCUUUCUCGCCUUGCCUCUCCUCGAC-CUGGGUGAAACGAGGG-GGGGAGGUGGAUG--------- -------------....(((......)))(..((((((((...(-((.(......).))))-)))))))..)...--------- ( -24.90, z-score = -1.01, R) >consensus ACUAGGCGAGAAAGCCCGGCCAUCUCGCCGUGUGUUAUUUGCCC_CCAAAUGGAGAAAUG__AACAGGCUGGAUGUGUG_____ ....(((......)))((((......))))....(((((((.....)))))))............................... (-10.12 = -9.17 + -0.96)



| Location | 20,953,569 – 20,953,645 |
|---|---|
| Length | 76 |
| Sequences | 9 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 68.37 |
| Shannon entropy | 0.64522 |
| G+C content | 0.54202 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -7.82 |
| Energy contribution | -7.59 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20953569 76 - 23011544 -----CACACAUCCAGCCUGUU--CAUUUCUCCAUUUGG-GGGCAAAUAACACACGGCGAGAUGGCCGGGCUUUCUCGCCUAGU -----....((((..(((((((--.((((((((.....)-))).))))))))...)))..))))((..(((......)))..)) ( -23.70, z-score = -1.78, R) >droSim1.chr2L_random 844034 76 - 909653 -----CACACAUCCAGACUGUU--CAUUUCUCCAUUUGG-GGGCAAAUAACACACGGCGAGAUGGCCGGGCUUUCUCGCCUAGU -----.............((((--.((((((((.....)-))).))))))))...(((((((.(((...))).))))))).... ( -22.60, z-score = -1.83, R) >droSec1.super_18 886654 76 - 1342155 -----CACACAUCCAGCCUGUU--CAUUUCUCCAUUUGG-GGGCAAAUAACACACGGCGAGAUGGCCGGGCUUUCUCGCCUAGU -----....((((..(((((((--.((((((((.....)-))).))))))))...)))..))))((..(((......)))..)) ( -23.70, z-score = -1.78, R) >droYak2.chr2R 19339237 76 + 21139217 -----CGCACUUCCGGCCUGUU--CAUUUCUCCAUUUGG-GGGCAAAUAACACACGGCGAGAUGGCCGGGCUUUCUCGCCUAGU -----.............((((--.((((((((.....)-))).))))))))...(((((((.(((...))).))))))).... ( -22.60, z-score = -0.39, R) >droEre2.scaffold_4845 19071388 74 + 22589142 -----CGCGCAGCCGGCCCGUU--CAUUUCUCCAUUUGG-GGGCAAGUAACACACGGC--GAUGGCCGGGCUUUCUCGCCUAGC -----.(((.((..((((((((--.((((((((.....)-))).)))))))....(((--....))))))))..)))))..... ( -26.70, z-score = -0.56, R) >droAna3.scaffold_12943 1268576 78 + 5039921 ---GACUUUCA-CGGCUCUGUU--CAUUUCUCCAUUUGGUGUGCAAAUAACACACGACUAGAUGGUCGGGCUUUCUCGCCUAGU ---........-.(((......--.......((((((((((((.........))).)))))))))..(((....)))))).... ( -17.30, z-score = -0.64, R) >dp4.chr4_group1 88408 84 + 5278887 UUCGAAGGUCCUUCUGCUUGUUGCCAUUUCUCCAUUUGGUGUGCAAAUAGCAGACCAAGAGAUGGCGAGGCGAGAAAGCCUAGU .....((((..(((((((..(((((((((((.....((((.(((.....))).)))))))))))))))))).)))).))))... ( -34.00, z-score = -3.90, R) >droPer1.super_5 26860 84 - 6813705 UUCGAAAGUCCUUCUGCUUGUUGCCAUUUCUCCAUUUGGUGUGCAAAUAGCAGACCAAGAGAUGGCGAGGCGAGAAAGCCUAGU ......((.(.(((((((..(((((((((((.....((((.(((.....))).)))))))))))))))))).)))).).))... ( -29.90, z-score = -3.03, R) >droWil1.scaffold_180772 3823017 60 + 8906247 ---------CAUCCACCUCCCC-CCCUCGUUUCACCCAG-GUCGAGGAGAGGCAAGGCGAGAAAGCCUAGC------------- ---------......((((...-.(((((..((.....)-).))))).))))..((((......))))...------------- ( -17.10, z-score = -0.89, R) >consensus _____CACACAUCCAGCCUGUU__CAUUUCUCCAUUUGG_GGGCAAAUAACACACGGCGAGAUGGCCGGGCUUUCUCGCCUAGU ..................((((.........((....)).........))))...(((......)))((((......))))... ( -7.82 = -7.59 + -0.23)
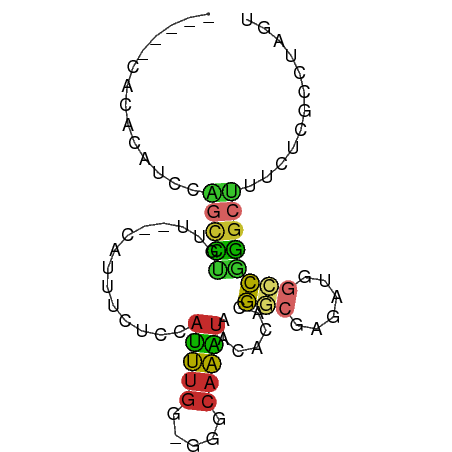
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:38 2011