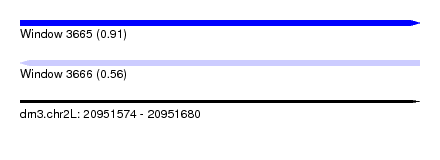
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,951,574 – 20,951,680 |
| Length | 106 |
| Max. P | 0.912273 |

| Location | 20,951,574 – 20,951,680 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.30635 |
| G+C content | 0.58351 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -35.33 |
| Energy contribution | -35.63 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
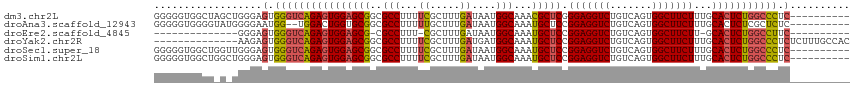
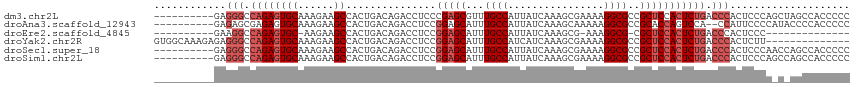
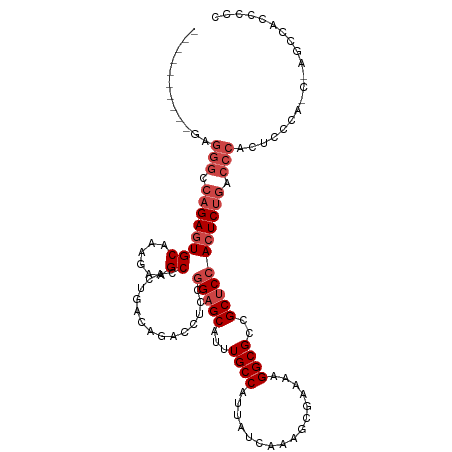
>dm3.chr2L 20951574 106 + 23011544 GGGGGUGGCUAGCUGGGAGUGGGUCAGAGUGGAGCGGCGCCUUUUCGCUUUGAUAAUGGCAAACGCUCGGGAGGUCUGUCAGUGGCUUCUUUGCACUCUGGCCCUC---------- ...(((.....)))......((((((((((((((((..(((...((.....))....)))...)))))((((((((.......))))))))..)))))))))))..---------- ( -43.20, z-score = -0.96, R) >droAna3.scaffold_12943 1266422 104 - 5039921 GGGGGUGGGGUAUGGGGAAUGG--UGGACUGGUGCGGCGCCUUUUUGCUUUGAUAAUGGCAAAUGCUCCGGAGGUCUGUCAGUGGCUUCUUUGCACUCUCGCUCUC---------- (((((((((((.(..(((((((--..((((....(((.((...((((((........)))))).)).)))..))))..))).....))))..).)).)))))))))---------- ( -38.50, z-score = -1.69, R) >droEre2.scaffold_4845 19069858 89 - 22589142 --------------GGGAGUGGGUCAGAGUGGAGCG-CGCCUUU-CGCUUUGAUAAUGGCAAAUGCUCCGGAGGUCUGUCAGUGGCUUCUU-GCACUCUGGCCUUC---------- --------------......((((((((((((((((-.(((..(-(.....))....)))...))))))(((((((.......))))))).-..))))))))))..---------- ( -36.40, z-score = -1.84, R) >droYak2.chr2R 19337477 102 - 21139217 --------------AAGAGUGGGUCAGAGUGGAGCGGCGCCUUUUCGCUUUGAUGAUGGCAAAUGCUCCGGAGGUCUGUCAGUGGCUUCUUUGCACUCUGGCCCUCUCUUUGCCAC --------------(((((.((((((((((((((((..(((...(((......))).)))...))))))(((((((.......)))))))....)))))))))).)))))...... ( -47.60, z-score = -3.70, R) >droSec1.super_18 884662 106 + 1342155 GGGGGUGGCUGGUUGGGAGUGGGUCAGAGUGGAGCGGCGCCUUUUCGCUUUGAUAAUGGCAAAUGCUCCGGAGGUCUGUCAGUGGCUUCUUUGCACUCUGGCCCUC---------- ....................((((((((((((((((..(((...((.....))....)))...))))))(((((((.......)))))))....))))))))))..---------- ( -40.80, z-score = -0.57, R) >droSim1.chr2L 20564745 106 + 22036055 GGGGGUGGCUGGCUGGGAGUGGGUCAGAGUGGAGCGGCGCCUUUUCGCUUUGAUAAUGGCAAAUGCUCCGGAGGUCUGUCAGUGGCUUCUUUGCACUCUGGCCCUC---------- ((((((((((.((.....)).))))(((((((((((..(((...((.....))....)))...))))))(((((((.......)))))))....))))).))))))---------- ( -42.90, z-score = -0.76, R) >consensus GGGGGUGGCU_G_UGGGAGUGGGUCAGAGUGGAGCGGCGCCUUUUCGCUUUGAUAAUGGCAAAUGCUCCGGAGGUCUGUCAGUGGCUUCUUUGCACUCUGGCCCUC__________ ....................((((((((((((((((..(((...((.....))....)))...))))).(((((((.......)))))))...)))))))))))............ (-35.33 = -35.63 + 0.31)
| Location | 20,951,574 – 20,951,680 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.30635 |
| G+C content | 0.58351 |
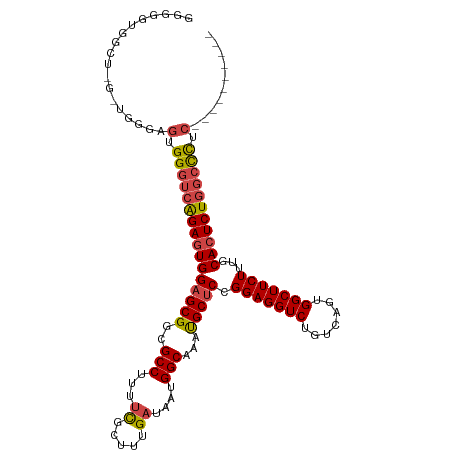
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -21.12 |
| Energy contribution | -22.46 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20951574 106 - 23011544 ----------GAGGGCCAGAGUGCAAAGAAGCCACUGACAGACCUCCCGAGCGUUUGCCAUUAUCAAAGCGAAAAGGCGCCGCUCCACUCUGACCCACUCCCAGCUAGCCACCCCC ----------..(((.(((((((...((......))............(((((..((((.((.((.....)).)))))).)))))))))))).))).................... ( -27.80, z-score = -1.17, R) >droAna3.scaffold_12943 1266422 104 + 5039921 ----------GAGAGCGAGAGUGCAAAGAAGCCACUGACAGACCUCCGGAGCAUUUGCCAUUAUCAAAGCAAAAAGGCGCCGCACCAGUCCA--CCAUUCCCCAUACCCCACCCCC ----------(((..(...(((((......).))))....)..)))(((.((.(((((..........)))))...)).)))..........--...................... ( -14.70, z-score = 0.32, R) >droEre2.scaffold_4845 19069858 89 + 22589142 ----------GAAGGCCAGAGUGC-AAGAAGCCACUGACAGACCUCCGGAGCAUUUGCCAUUAUCAAAGCG-AAAGGCG-CGCUCCACUCUGACCCACUCCC-------------- ----------...((.((((((((-.....))...............(((((.((((.......))))(((-.....))-)))))))))))).)).......-------------- ( -23.90, z-score = -1.06, R) >droYak2.chr2R 19337477 102 + 21139217 GUGGCAAAGAGAGGGCCAGAGUGCAAAGAAGCCACUGACAGACCUCCGGAGCAUUUGCCAUCAUCAAAGCGAAAAGGCGCCGCUCCACUCUGACCCACUCUU-------------- ......(((((.(((.((((((((......))...............(((((...((((.((........))...))))..))))))))))).))).)))))-------------- ( -33.40, z-score = -1.55, R) >droSec1.super_18 884662 106 - 1342155 ----------GAGGGCCAGAGUGCAAAGAAGCCACUGACAGACCUCCGGAGCAUUUGCCAUUAUCAAAGCGAAAAGGCGCCGCUCCACUCUGACCCACUCCCAACCAGCCACCCCC ----------..(((.((((((((......))...............(((((...((((.((.((.....)).))))))..))))))))))).))).................... ( -29.30, z-score = -2.03, R) >droSim1.chr2L 20564745 106 - 22036055 ----------GAGGGCCAGAGUGCAAAGAAGCCACUGACAGACCUCCGGAGCAUUUGCCAUUAUCAAAGCGAAAAGGCGCCGCUCCACUCUGACCCACUCCCAGCCAGCCACCCCC ----------..(((.((((((((......))...............(((((...((((.((.((.....)).))))))..))))))))))).))).................... ( -29.30, z-score = -1.47, R) >consensus __________GAGGGCCAGAGUGCAAAGAAGCCACUGACAGACCUCCGGAGCAUUUGCCAUUAUCAAAGCGAAAAGGCGCCGCUCCACUCUGACCCACUCCCA_C_AGCCACCCCC ............(((.((((((((......))...............(((((...((((................))))..))))))))))).))).................... (-21.12 = -22.46 + 1.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:37 2011