
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,951,082 – 20,951,179 |
| Length | 97 |
| Max. P | 0.730402 |

| Location | 20,951,082 – 20,951,179 |
|---|---|
| Length | 97 |
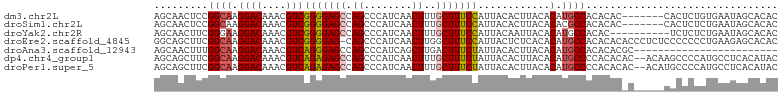
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Shannon entropy | 0.40830 |
| G+C content | 0.51896 |
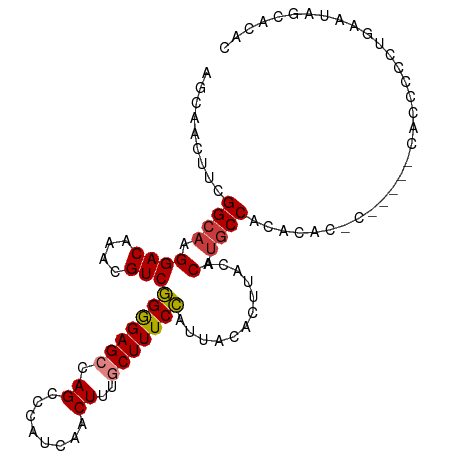
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -14.16 |
| Energy contribution | -14.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20951082 97 - 23011544 AGCAACUCCGGCAAGGACAAACGUCGGGGAGCCAGCCCAUCAACUUUGCUUUCCAUUACACUUACACAUGCCACACAC-------CACUCUGUGAAUAGCACAC .((......((((.((((....)))((..(((.((........))..)))..))............).)))).((((.-------.....))))....)).... ( -23.40, z-score = -2.01, R) >droSim1.chr2L 20564253 97 - 22036055 AGCAACUCCGGCAAGGACAAACGUCGGGGAGCCAGCCCAUCAACUUUGCUUUCCAUUACACUUACACACGCCACACAC-------CACUCUCUGAAUAGCACAC .((......(((...(((....)))((..(((.((........))..)))..))...............)))...((.-------.......))....)).... ( -18.10, z-score = -1.04, R) >droYak2.chr2R 19337008 94 + 21139217 AGCAACUUCGGGAAGGACAAACGUCGGGGAGCCAGCCCAUCAACUUUGCUUUCCAUUACAAUUACACAUGCCACAC----------UCUCUCUGAAUAGCACAC .((...(((((..(((((....)))((..(((.((........))..)))..))......................----------.))..)))))..)).... ( -20.20, z-score = -1.26, R) >droEre2.scaffold_4845 19069433 103 + 22589142 GGCAGCUUCGGCAAGGACAAACGUCGGGGAG-CAGCCCAUCAACUUGGCUUUCCAUUACUCUCACACAUGCCACACACACCCUCUCCCCCCCUGAAGAGCACAC .((..((((((....(((....)))((((((-.((..........((((....................))))........))))))))..)))))).)).... ( -25.12, z-score = -0.73, R) >droAna3.scaffold_12943 1266152 80 + 5039921 AGCAACUUUGGCAAGGACAAACGUCAGGGAGCCAGCCCAUCAGCUUGACUUUCUAUUACACUUACACAUGCCACACACGC------------------------ .((.....(((((.((((....)))((..(((.(((......))).).))..))............).))))).....))------------------------ ( -17.40, z-score = -1.31, R) >dp4.chr4_group1 85708 102 + 5278887 AGCAGCUUCGGCAAGGACAAACGUCAGAGAGCCAGCCCAUCAACUUUGCUUUCUAUUACACUUACACAUGCCCCACACAC--ACAAGCCCCAUGCCUCACAUAC .(((((....))((((((....)))(((((((.((........))..)))))))......))).....))).........--...................... ( -16.90, z-score = -1.08, R) >droPer1.super_5 24132 102 - 6813705 AGCAGCUUCGGCAAGGACAAACGUCAGAGAGCCAGCCCAUCAACUUUGCUUUCUAUUACACUUACACAUGCCCCACACAC--ACAUGCCCCAUGCCUCACAUAC .(((.....((((.((((....)))(((((((.((........))..)))))))..........................--.).))))...)))......... ( -17.70, z-score = -1.13, R) >consensus AGCAACUUCGGCAAGGACAAACGUCGGGGAGCCAGCCCAUCAACUUUGCUUUCCAUUACACUUACACAUGCCACACAC_C_____CACCCCCUGAAUAGCACAC .........((((.((((....)))(((((((.((........))..)))))))............).))))................................ (-14.16 = -14.04 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:35 2011