
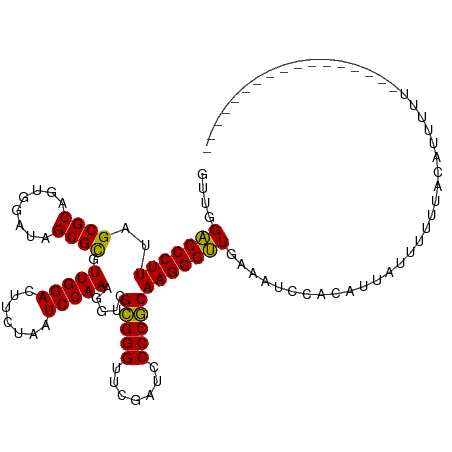
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,965,421 – 1,965,525 |
| Length | 104 |
| Max. P | 0.998519 |

| Location | 1,965,421 – 1,965,525 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Shannon entropy | 0.54482 |
| G+C content | 0.47863 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.18 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1965421 104 + 23011544 GUUGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAAGUACUCCUUUUUUUGUACAUGCUAA---------------- .((.(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).))((((..........)))).......---------------- ( -33.00, z-score = -1.43, R) >droPer1.super_1 10184725 104 + 10282868 GUUAGACCCUUUGGCGCAGCGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGCAACCAAAAUCACAUUUUAUAUUCUAA---------------- .(((((.((((((((((........))))(((((....))))).)))))).....(((((((..(((....)))))).)))).................)))))---------------- ( -30.10, z-score = -1.04, R) >dp4.chr4_group3 8732046 104 + 11692001 GUUAGACCCUUUGGCGCAGCGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGCAACCAAAAUCACAUUUUAUAUUCUAA---------------- .(((((.((((((((((........))))(((((....))))).)))))).....(((((((..(((....)))))).)))).................)))))---------------- ( -30.10, z-score = -1.04, R) >droSim1.chr2L 1935051 105 + 22036055 GUUGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAAGUACUCCUUUUUUUGUACAUGCUUAG--------------- .((.(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).))((((..........))))........--------------- ( -33.00, z-score = -1.43, R) >droSec1.super_14 1908573 98 + 2068291 GUUGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAAGUACUCCUUUUUUUGUUUA---------------------- .((.(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).))...................---------------------- ( -29.90, z-score = -1.01, R) >droYak2.chr2L 1946731 102 + 22324452 GUAGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAUUGCCACUAGAUGUACUACUUUUU------------------ ((((.((......((((........)))).(((((.......))))).((((((((..((((..(((....)))))))))))))))....)).)))).....------------------ ( -33.80, z-score = -1.82, R) >droEre2.scaffold_4929 2011441 110 + 26641161 GUAGGACCCUUUAGCGCAUUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUGGCGGGUUCGAUUCCCGCAAGGGUUGAUUGCCAUUAGCUGUGCUACUUUUUUGGCAAAG---------- ....(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))..((((((.((((...))))......))))))..---------- ( -36.80, z-score = -1.31, R) >droWil1.scaffold_180772 1186091 119 - 8906247 GUACGACCCUUUAGCGCAGAGGAUAGCGUGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGCAUAUCAGAUGAGACUUGUAAUUUUUUUAUUAAACAAAUCAG- ....((((.....((((........)))).(((((.......))))).))))(((((.......)))))((((((((((..((......))..))))))))))................- ( -31.30, z-score = -1.14, R) >droAna3.scaffold_12916 9677443 111 - 16180835 GUAAGACCCUUUGGCGCAGCGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGAAAAC-ACAAACUGUGUUACUUAUAUUUAAAAGAU-------- (((((((((((.(((((.((.....))))))).(((((.........)))))(((((.......)))))))))))....(((-((.....))))).)))))...........-------- ( -33.80, z-score = -1.73, R) >droVir3.scaffold_12963 7560759 120 - 20206255 AUUAGACCCUUUGGCGCAGCGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGUUAUUAGAAUUACUUUUUUUAUUUUUCAAAAAUUGUUCUAUAC ....(((((((.(((((.((.....))))))).(((((.........)))))(((((.......)))))))))))).....((((((....(((((........)))))..))))))... ( -32.10, z-score = -1.54, R) >droMoj3.scaffold_6500 7423555 103 + 32352404 GUUCGACCCUUUAGCGCAGUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUUCCCGCAAGGGUUGUAUUUCUUAUUUUUUAGAGUAUUUUU----------------- ...((((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))))...((((........)))).......----------------- ( -31.10, z-score = -2.13, R) >anoGam1.chr2R 49513013 91 + 62725911 GUGUGUCCCUUUGGCGCAGCGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGUGGGUUCGAUCCCCACAAGGGAUGACGGUUACGUUCUU----------------------------- ....(((((((.(((((.((.....))))))).(((((.........)))))(((((.......))))))))))))((((....))))...----------------------------- ( -29.80, z-score = -0.46, R) >consensus GUUGGACCCUUUAGCGCAGUGGAUAGCGCGUUGGACUUCUAAUCCAAAGGUCGCGGGUUCGAUCCCCGCAAGGGUUGAAAUCCACAUUAUUUUUUACAUUUUU_________________ ....(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))............................................ (-28.56 = -28.18 + -0.38)

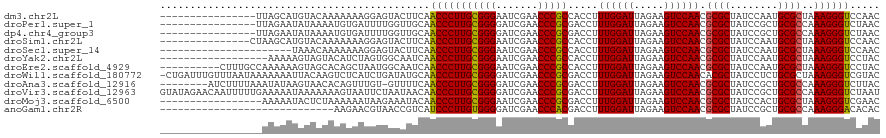
| Location | 1,965,421 – 1,965,525 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Shannon entropy | 0.54482 |
| G+C content | 0.47863 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.48 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1965421 104 - 23011544 ----------------UUAGCAUGUACAAAAAAAGGAGUACUUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAAC ----------------.......((((..........))))....(((((((((((.......))))).....((((((.....)))))).((((........))))..))))))..... ( -29.90, z-score = -2.35, R) >droPer1.super_1 10184725 104 - 10282868 ----------------UUAGAAUAUAAAAUGUGAUUUUGGUUGCAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAAGGGUCUAAC ----------------(((((........((..((....))..)).((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))). ( -34.40, z-score = -2.39, R) >dp4.chr4_group3 8732046 104 - 11692001 ----------------UUAGAAUAUAAAAUGUGAUUUUGGUUGCAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAAGGGUCUAAC ----------------(((((........((..((....))..)).((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))). ( -34.40, z-score = -2.39, R) >droSim1.chr2L 1935051 105 - 22036055 ---------------CUAAGCAUGUACAAAAAAAGGAGUACUUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAAC ---------------........((((..........))))....(((((((((((.......))))).....((((((.....)))))).((((........))))..))))))..... ( -29.90, z-score = -2.33, R) >droSec1.super_14 1908573 98 - 2068291 ----------------------UAAACAAAAAAAGGAGUACUUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCAAC ----------------------............(((....))).(((((((((((.......))))).....((((((.....)))))).((((........))))..))))))..... ( -28.30, z-score = -2.63, R) >droYak2.chr2L 1946731 102 - 22324452 ------------------AAAAAGUAGUACAUCUAGUGGCAAUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCUAC ------------------.....(((((((.....))).......(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))).)))) ( -29.70, z-score = -2.14, R) >droEre2.scaffold_4929 2011441 110 - 26641161 ----------CUUUGCCAAAAAAGUAGCACAGCUAAUGGCAAUCAACCCUUGCGGGAAUCGAACCCGCCACCUUUGGAUUAGAAGUCCAACGCGCUAUCCAAUGCGCUAAAGGGUCCUAC ----------..((((((......((((...)))).))))))...(((((((((((.......))))).....((((((.....)))))).((((........))))..))))))..... ( -36.20, z-score = -3.29, R) >droWil1.scaffold_180772 1186091 119 + 8906247 -CUGAUUUGUUUAAUAAAAAAAUUACAAGUCUCAUCUGAUAUGCAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACACGCUAUCCUCUGCGCUAAAGGGUCGUAC -..(((((((..............)))))))..........(((.(((((((((((.......))))).....((((((.....))))))..(((........)))...)))))).))). ( -33.74, z-score = -2.75, R) >droAna3.scaffold_12916 9677443 111 + 16180835 --------AUCUUUUAAAUAUAAGUAACACAGUUUGU-GUUUUCAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAAGGGUCUUAC --------............((((.(((((.....))-)))....(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))))). ( -33.90, z-score = -2.50, R) >droVir3.scaffold_12963 7560759 120 + 20206255 GUAUAGAACAAUUUUUGAAAAAUAAAAAAAGUAAUUCUAAUAACAACCCUUGCGGGGAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAAGGGUCUAAU ...(((((..((((((..........))))))..)))))............(((((.......)))))(((((((((((.....))))...((((........))))..))))))).... ( -34.40, z-score = -3.25, R) >droMoj3.scaffold_6500 7423555 103 - 32352404 -----------------AAAAAUACUCUAAAAAAUAAGAAAUACAACCCUUGCGGGAAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCACUGCGCUAAAGGGUCGAAC -----------------..................................(((((.......)))))(((((((((((.....))))...((((........))))..))))))).... ( -29.60, z-score = -3.68, R) >anoGam1.chr2R 49513013 91 - 62725911 -----------------------------AAGAACGUAACCGUCAUCCCUUGUGGGGAUCGAACCCACGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCGCUGCGCCAAAGGGACACAC -----------------------------..((.((....)))).(((((((((((.......))))).....((((((.....)))))).((((........))))..))))))..... ( -29.40, z-score = -2.02, R) >consensus _________________AAAAAUAUAAAAAAUAAUGUGGAAUUCAACCCUUGCGGGAAUCGAACCCGCGACCUUUGGAUUAGAAGUCCAACGCGCUAUCCACUGCGCUAAAGGGUCCAAC .............................................(((((((((((.......))))).....((((((.....)))))).((((........))))..))))))..... (-28.70 = -28.48 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:44 2011