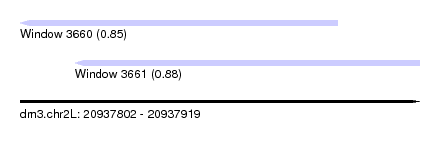
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,937,802 – 20,937,919 |
| Length | 117 |
| Max. P | 0.877052 |

| Location | 20,937,802 – 20,937,895 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.55141 |
| G+C content | 0.35279 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -10.87 |
| Energy contribution | -10.58 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20937802 93 - 23011544 -UGAUCGGGCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUA--AUAUG-AGUUCCCAUU---------------- -...(((((((.....)))))..(((((((((((((((((.............)))).)))))))))...)))).....--....)-).........---------------- ( -20.12, z-score = -2.32, R) >droAna3.scaffold_12943 1247960 94 + 5039921 GGGUGGGCCGUGGAAAGCCUAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACAGUAAUUAAGUGAUAUCAUGCUAUCUA--AUAUG-AGCUCCCAUU---------------- ((((((((........)))))..(((.((((((((((((((...........))))).)))))))))(((((.......--.))))-)))))))...---------------- ( -23.30, z-score = -1.34, R) >droEre2.scaffold_4845 19056600 93 + 22589142 -UGAUCGGCCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCGUGCUAUCUA--AUACG-AGUUCCCCCG---------------- -...(((((........)))...(((((((((((((((((.............)))).))))))))...))))).....--....)-).........---------------- ( -15.02, z-score = -1.33, R) >droYak2.chr2R 19323838 93 + 21139217 -UGAUCGGUCUCUCAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUA--AUAUG-AGCUCCCAUU---------------- -.....((...((((........(((((((((((((((((.............)))).)))))))))...)))).....--...))-))...))...---------------- ( -16.31, z-score = -1.48, R) >droSec1.super_18 869758 93 - 1342155 -UGAUCGGGCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUA--AUAUG-AGUUCCCAUU---------------- -...(((((((.....)))))..(((((((((((((((((.............)))).)))))))))...)))).....--....)-).........---------------- ( -20.12, z-score = -2.32, R) >droSim1.chr2L 20550888 93 - 22036055 -UGAUCGGGCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUA--AUAUG-AGUUCCCAUU---------------- -...(((((((.....)))))..(((((((((((((((((.............)))).)))))))))...)))).....--....)-).........---------------- ( -20.12, z-score = -2.32, R) >droWil1.scaffold_180772 3797450 105 + 8906247 -AGUGAGGCAUCUAAAUUCAAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAGCAUGCUAUCUA-UAGUUAUUGUUGGUUUUUUUU--UGCUUU---- -...((((((...(((..(((...((.(((((((((((((.............)))).)))))))))))..((((....-)))).....)))..)))....--))))))---- ( -18.52, z-score = -0.20, R) >droPer1.super_5 9074 111 - 6813705 -UGGGGGCUUUCGAUAUGUUAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUACUAUAUG-AGUCCGUGUCCGUCCCCAUCGCCAAC -(((((((....((((((.....(((((((((((((((((.............)))).)))))))))...)))).((........)-)...)))))).)))))))........ ( -27.42, z-score = -2.22, R) >dp4.chr4_group1 70572 111 + 5278887 -UGGGGGCUUUCGAUAUGUUAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUACUAUAUG-AGUCCGUGUCCGUCCCCAUCGCCAAC -(((((((....((((((.....(((((((((((((((((.............)))).)))))))))...)))).((........)-)...)))))).)))))))........ ( -27.42, z-score = -2.22, R) >droVir3.scaffold_12963 9506109 107 + 20206255 -UGCAAAGCUUCGAAUUUC---UAUCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAGCAUGGUAUCUA-UUUAUA-UGCUGGCUGUUGUUGCUGCUGUUGUU -.(((((((..(((....(---..((((((((((((((((.............)))).)))))))))(((((.((....-..))))-))))))..)...)))..))).)))). ( -18.62, z-score = 0.75, R) >droGri2.scaffold_15252 8464303 107 + 17193109 -UGAAAAGCAUCGAAUUUC---UAUCAUGUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAAUGCAUGGUAUCUA-UUUAUG-UGCUGAUUGUUGUUGUUACUGUUGUU -.....(((((.((((...---((((((((((((((((((.............)))).))))))...))))))))...)-)))..)-))))...................... ( -17.32, z-score = 0.42, R) >consensus _UGAUCGGCCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCUA__AUAUG_AGUUCCCAUU________________ .......................(((((((((((((((((.............)))).))))))))...)))))....................................... (-10.87 = -10.58 + -0.29)


| Location | 20,937,818 – 20,937,919 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.17 |
| Shannon entropy | 0.58446 |
| G+C content | 0.36400 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -10.84 |
| Energy contribution | -10.55 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20937818 101 - 23011544 --------------CACACAUUCAUUCGGGGCUAUUCCUGAUCGGGCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU --------------...........((((((....))))))..(((((.....)))))..(((((((((((((((((.............)))).)))))))))...)))).... ( -25.62, z-score = -3.11, R) >droAna3.scaffold_12943 1247976 115 + 5039921 GAUAUAUUCGUUUGAGGUUUUUCGAUAGCGGCCAAGUGGGUGGGCCGUGGAAAGCCUAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACAGUAAUUAAGUGAUAUCAUGCUAUCU ((((.....((((.(((((((((....((((((.........)))))))))))))))..))))(((((((((((((((...........))))).)))))))))).....)))). ( -32.70, z-score = -2.41, R) >droEre2.scaffold_4845 19056616 88 + 22589142 ---------------------------GGGGCCAUUUCUGAUCGGCCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCGUGCUAUCU ---------------------------((((((..........))))))...........(((((((((((((((((.............)))).))))))))...))))).... ( -21.22, z-score = -1.96, R) >droYak2.chr2R 19323854 101 + 21139217 --------------CCCACUUUCAUUCGGGUUUAUUUGUGAUCGGUCUCUCAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU --------------.............((((...((((.((......)).))))))))..(((((((((((((((((.............)))).)))))))))...)))).... ( -19.72, z-score = -2.04, R) >droSec1.super_18 869774 101 - 1342155 --------------CACACAUUCAUUCGGGGCUAUUUCUGAUCGGGCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU --------------...........((((((....))))))..(((((.....)))))..(((((((((((((((((.............)))).)))))))))...)))).... ( -22.82, z-score = -2.29, R) >droSim1.chr2L 20550904 101 - 22036055 --------------CACACAUUCAUUCGGGGCUAUUUCUGAUCGGGCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU --------------...........((((((....))))))..(((((.....)))))..(((((((((((((((((.............)))).)))))))))...)))).... ( -22.82, z-score = -2.29, R) >droWil1.scaffold_180772 3797478 88 + 8906247 ---------------------------GGGAGGAAUGUAGUGAGGCAUCUAAAUUCAAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAGCAUGCUAUCU ---------------------------.(((.((((.(((........))).))))....(((((((((((((((((.............)))).)))))))))...)))).))) ( -15.42, z-score = 0.05, R) >droPer1.super_5 9108 92 - 6813705 -----------------------AAUUGAGCUACAUUUUGGGGGCUUUCGAUAUGUUAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU -----------------------...(((((.(((...(((..(((((..(....)..)))))...)))..))).)))))....................((((......)))). ( -17.70, z-score = -0.78, R) >dp4.chr4_group1 70606 92 + 5278887 -----------------------AAUGGAGCUACAUUUUGGGGGCUUUCGAUAUGUUAAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU -----------------------....((((.(((...(((..(((((..(....)..)))))...)))..))).)))).....................((((......)))). ( -17.00, z-score = -0.32, R) >droVir3.scaffold_12963 9506142 100 + 20206255 -----------UGUGCG-GCUGUGUGCGUGUGUGUGUGUGCAAAGCUUCGAAUUUCU---AUCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAGCAUGGUAUCU -----------.(((((-(((...((((..(....)..)))).))))..........---....(((((((((((((.............)))).)))))))))))))....... ( -22.32, z-score = 0.03, R) >droGri2.scaffold_15252 8464336 101 + 17193109 -----------UAUAAGCGUGGUAAAUGUGUGCGUGUGUGAAAAGCAUCGAAUUUCU---AUCAUGUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAAUGCAUGGUAUCU -----------.....((((..((((((...(((((...((((.........)))).---..)))))))))))..))((((((((.........))))))))..))......... ( -18.70, z-score = 0.25, R) >consensus ____________________UUCAUUCGGGGCUAUUUCUGAUCGGCCUCUAAAAGCCCAAAGCAUAUCAUUUGUUGCUCAUUUAAAUACCGUAAUUAAGUGAUAUCAUGCUAUCU ............................................................(((((((((((((((((.............)))).))))))))...))))).... (-10.84 = -10.55 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:33 2011