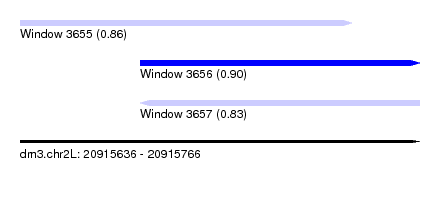
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,915,636 – 20,915,766 |
| Length | 130 |
| Max. P | 0.904059 |

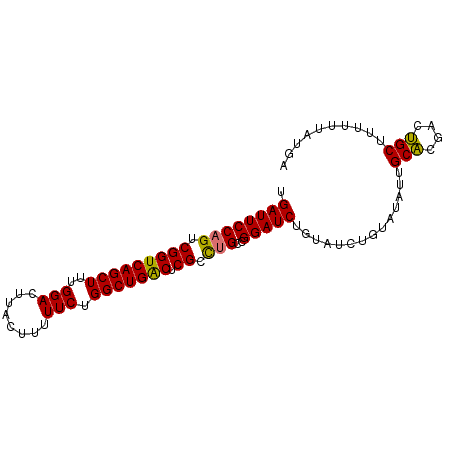
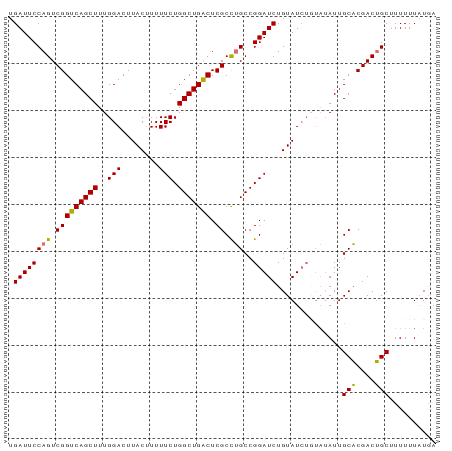
| Location | 20,915,636 – 20,915,744 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.40 |
| Shannon entropy | 0.55557 |
| G+C content | 0.43106 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -12.23 |
| Energy contribution | -14.97 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.863147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
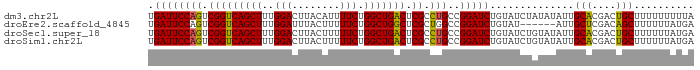
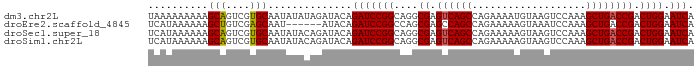
>dm3.chr2L 20915636 108 + 23011544 UGUGCAACGCGACGUUGACUUAUUGAAAUCGAUUGAUUCUGAUUCCAGUCGGUCAGCUUUGGACU------UACAUUUU-----CUGGCUGACUCGCCUGCCGGAUCUGUAUCUAUAUA .............(((((((.((((.((((((.....)).)))).)))).)))))))..((((..------((((...(-----(((((.(......).))))))..)))))))).... ( -28.50, z-score = -1.48, R) >droAna3.scaffold_12943 1219981 109 - 5039921 ----UAAGCUUAUGGUGACUUAUUGAAAUCGAUUGAUUCUGAUUCCAGUCUGUCGGCUUUGGUUUCUAUUCCAUUUUUUUUACCCUAGACGACUUGCCUGCCGGAUCUGUAAA------ ----...((...(((((((..((((.((((((.....)).)))).))))..)))(((...((((((((.................)))).)))).))).)))).....))...------ ( -20.73, z-score = -0.30, R) >droEre2.scaffold_4845 19038025 102 - 22589142 UGUGCAACGCGACGUUGACUUAUUGAAAUCGAUUGAUUCUGAUUCCAGUCGGUCAGCUUUGGAUU------UACUUUUU-----CUGGCUGGCUCGCUGGCCGGAUCUGUAUA------ ((((((...(((.(((((((.((((.((((((.....)).)))).)))).))))))).)))....------.......(-----(((((..(....)..))))))..))))))------ ( -34.40, z-score = -3.18, R) >droYak2.chr2R 19304075 102 - 21139217 UGUGCAACGCGACGUUGACUUAUUGAAAUCGAUUGAUUCUGAUUUCAUUCGGUCAGCUUUGGAUU------UACUUUUU-----CUGGCUGGUUCGCUGGCCGGAUCUGUAUA------ ((((((...(((.(((((((...(((((((((.....)).)))))))...))))))).)))....------.......(-----(((((..(....)..))))))..))))))------ ( -35.60, z-score = -4.35, R) >droSec1.super_18 849398 108 + 1342155 UGUGCAACGCGACGUUGACUUAUUAAAAUCGAUUGAUUCUGAUUCCAGUCGGUCAGCUUUGGACU------UACUUUUU-----CUGGCUGACUCGCCUGCCGGAUCUGUAUCUGUAUA ((((((...(((..((((....))))..)))...(((...((((((((.(((((((((..(((..------......))-----).))))))).)).)))..)))))...))))))))) ( -28.10, z-score = -1.57, R) >droSim1.chr2L 20530877 107 + 22036055 UGUGCAACGCGACGUUGACUUAUUAAA-UCGAUUGAUUCUGAUUCCAGUCGGUCAGCUUUGGACU------UACUUUUU-----CUGGCUGACUCGCCUGCCGGAUCUGUAUCUGUAUA ((((((...(((.(((((((.(((.((-((((.....)).))))..))).))))))).)))....------(((....(-----(((((.(......).))))))...)))..)))))) ( -26.40, z-score = -0.99, R) >droWil1.scaffold_180708 8934112 89 + 12563649 --------------------AAUUGAAAGCGACAUUGACUUAUUGCCGAUAUUCAGAUUGAUAUU----UCUGUCUGCUG----UCUGCAGUUGCAUUUGCCGGAUCUAUAAUCGUG-- --------------------........((((...((....(((((.((((..(((((.((....----)).))))).))----)).)))))..)).))))................-- ( -17.60, z-score = 0.05, R) >consensus UGUGCAACGCGACGUUGACUUAUUGAAAUCGAUUGAUUCUGAUUCCAGUCGGUCAGCUUUGGAUU______UACUUUUU_____CUGGCUGACUCGCCUGCCGGAUCUGUAUA__U___ .........(((.(((((((.((((.(((((........))))).)))).))))))).))).......................(((((.(......).)))))............... (-12.23 = -14.97 + 2.74)

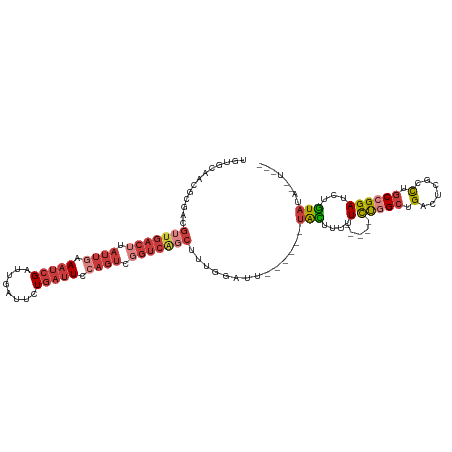
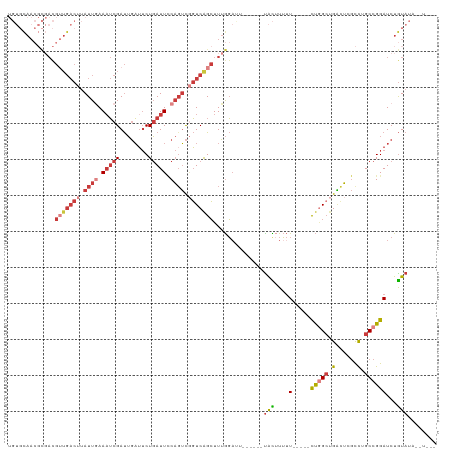
| Location | 20,915,675 – 20,915,766 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Shannon entropy | 0.14130 |
| G+C content | 0.44712 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -20.73 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20915675 91 + 23011544 UGAUUCCAGUCGGUCAGCUUUGGACUUACAUUUUCUGGCUGACUCGCCUGCCGGAUCUGUAUCUAUAUAUUGCACGACUGCUUUUUUUUUA ......((((((..(((...((((..((((...((((((.(......).))))))..))))))))....)))..))))))........... ( -23.50, z-score = -2.67, R) >droEre2.scaffold_4845 19038064 85 - 22589142 UGAUUCCAGUCGGUCAGCUUUGGAUUUACUUUUUCUGGCUGGCUCGCUGGCCGGAUCUGUAU------AUUGCUCGACAGCUUUUUUAUGA .(((((((((.((((((((..(((........))).)))))))).)))))...)))).....------...(((....))).......... ( -23.90, z-score = -1.39, R) >droSec1.super_18 849437 91 + 1342155 UGAUUCCAGUCGGUCAGCUUUGGACUUACUUUUUCUGGCUGACUCGCCUGCCGGAUCUGUAUCUGUAUAUUGCACGACUGCUUUUUUAUGA ......(((((((((((((..(((........))).)))))))..((....(((((....)))))......)).))))))........... ( -23.30, z-score = -1.90, R) >droSim1.chr2L 20530915 91 + 22036055 UGAUUCCAGUCGGUCAGCUUUGGACUUACUUUUUCUGGCUGACUCGCCUGCCGGAUCUGUAUCUGUAUAUUGCACGACUGCUUUUUUAUGA ......(((((((((((((..(((........))).)))))))..((....(((((....)))))......)).))))))........... ( -23.30, z-score = -1.90, R) >consensus UGAUUCCAGUCGGUCAGCUUUGGACUUACUUUUUCUGGCUGACUCGCCUGCCGGAUCUGUAUCUGUAUAUUGCACGACUGCUUUUUUAUGA .((((((((.(((((((((..(((........))).))))))).)).)))..)))))..............(((....))).......... (-20.73 = -20.23 + -0.50)
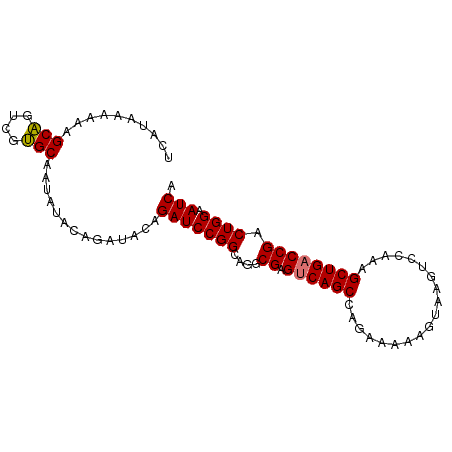
| Location | 20,915,675 – 20,915,766 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Shannon entropy | 0.14130 |
| G+C content | 0.44712 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.03 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20915675 91 - 23011544 UAAAAAAAAAGCAGUCGUGCAAUAUAUAGAUACAGAUCCGGCAGGCGAGUCAGCCAGAAAAUGUAAGUCCAAAGCUGACCGACUGGAAUCA ..........(((....)))..............(((((((....((.((((((...................)))))))).)))).))). ( -18.51, z-score = -1.32, R) >droEre2.scaffold_4845 19038064 85 + 22589142 UCAUAAAAAAGCUGUCGAGCAAU------AUACAGAUCCGGCCAGCGAGCCAGCCAGAAAAAGUAAAUCCAAAGCUGACCGACUGGAAUCA ..........(((....)))...------.....(((....((((((.(.((((...................)))).))).)))).))). ( -15.01, z-score = -0.45, R) >droSec1.super_18 849437 91 - 1342155 UCAUAAAAAAGCAGUCGUGCAAUAUACAGAUACAGAUCCGGCAGGCGAGUCAGCCAGAAAAAGUAAGUCCAAAGCUGACCGACUGGAAUCA ..........(((....)))..............(((((((....((.((((((...................)))))))).)))).))). ( -18.51, z-score = -1.17, R) >droSim1.chr2L 20530915 91 - 22036055 UCAUAAAAAAGCAGUCGUGCAAUAUACAGAUACAGAUCCGGCAGGCGAGUCAGCCAGAAAAAGUAAGUCCAAAGCUGACCGACUGGAAUCA ..........(((....)))..............(((((((....((.((((((...................)))))))).)))).))). ( -18.51, z-score = -1.17, R) >consensus UCAUAAAAAAGCAGUCGUGCAAUAUACAGAUACAGAUCCGGCAGGCGAGUCAGCCAGAAAAAGUAAGUCCAAAGCUGACCGACUGGAAUCA ..........(((....)))..............(((((((....((.((((((...................)))))))).)))).))). (-17.61 = -17.48 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:30 2011