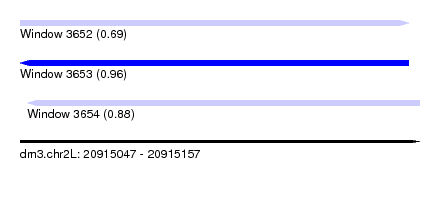
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,915,047 – 20,915,157 |
| Length | 110 |
| Max. P | 0.956377 |

| Location | 20,915,047 – 20,915,154 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.66 |
| Shannon entropy | 0.16649 |
| G+C content | 0.43186 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -24.14 |
| Energy contribution | -25.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
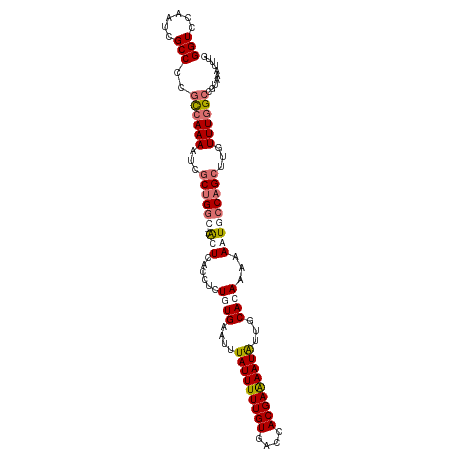
>dm3.chr2L 20915047 107 + 23011544 -GCCAAAUUUACGGCCAAACAAGCUGGCAUUUUUUGUGCAACAUUUUCGUGGUCACAAAAAUAAUUUCAAAGAGGUGAGCAAGCCAGCGAUUUUGGCGGGGCGAUUGG -.((((.....(.((((((...((((((..(((((((((.((......)).).))))))))....((((......))))...))))))...)))))).).....)))) ( -33.70, z-score = -2.28, R) >droEre2.scaffold_4845 19037441 104 - 22589142 GACUAAAUUUACGGC-AAACAAGCGGUUAUUUUUUGUGCAAUAUUCUCGUGGUCACAAAAAUAAAUUCAAAGAGGUGAGU--GCCAGCGAAUUUUA-GGGGCGAUUGG ..(((((.((.((((-......))(((...((((((((...(((....)))..))))))))...(((((......)))))--)))..)))).))))-).......... ( -16.00, z-score = 1.51, R) >droSec1.super_18 848825 104 + 1342155 --CCAAAUUUACGGCCAAACAAGCUGGAACUUUUUGUGCAAUAUUUUCGUGGUCACAAAAAUAAAUUCACAGAGGUGAGU--GCCAGUGAUUUUGGCGGGGCGAUUGG --((((.....(.((((((...(((((.((((((((((...((((((.((....)).))))))....))))))...))))--.)))))...)))))).).....)))) ( -30.90, z-score = -2.10, R) >droSim1.chr2L 20530300 104 + 22036055 --CCAAAUUUACGGCCAAACAAGCUGGCACUUUUUGUGCAAUAUUUUCGUGGUCACAAAAAUAAAUUCACAGAGGUGAGU--GCCAGCGAUUUUGGCGGGGCGAUUGG --((((.....(.((((((...((((((((((((((((...((((((.((....)).))))))....))))))...))))--))))))...)))))).).....)))) ( -39.10, z-score = -4.37, R) >consensus __CCAAAUUUACGGCCAAACAAGCUGGCACUUUUUGUGCAAUAUUUUCGUGGUCACAAAAAUAAAUUCAAAGAGGUGAGU__GCCAGCGAUUUUGGCGGGGCGAUUGG ..((((.....(.((((((...((((((..((((((((...(((....)))..))))))))...((((((....))))))..))))))...)))))).).....)))) (-24.14 = -25.70 + 1.56)

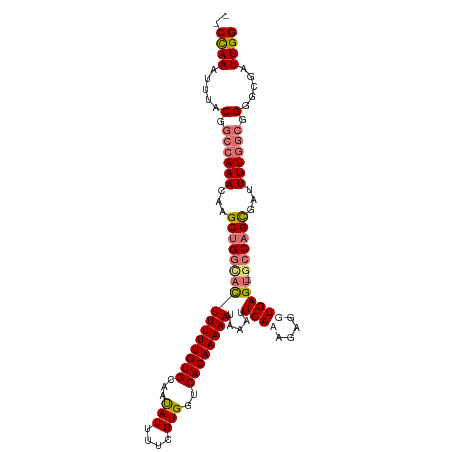
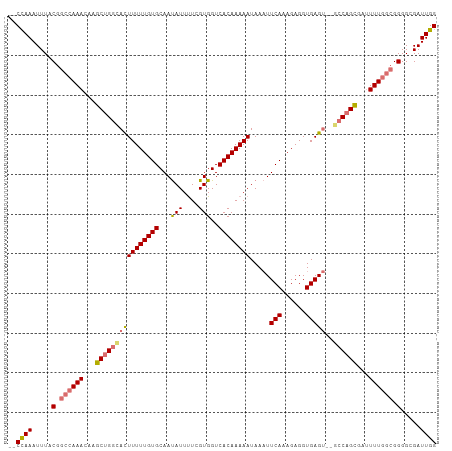
| Location | 20,915,047 – 20,915,154 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Shannon entropy | 0.16649 |
| G+C content | 0.43186 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -18.51 |
| Energy contribution | -19.95 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20915047 107 - 23011544 CCAAUCGCCCCGCCAAAAUCGCUGGCUUGCUCACCUCUUUGAAAUUAUUUUUGUGACCACGAAAAUGUUGCACAAAAAAUGCCAGCUUGUUUGGCCGUAAAUUUGGC- ((((.......((((((...((((((....(((......))).....((((((((.(.((......)).)))))))))..))))))...)))))).......)))).- ( -30.44, z-score = -2.84, R) >droEre2.scaffold_4845 19037441 104 + 22589142 CCAAUCGCCCC-UAAAAUUCGCUGGC--ACUCACCUCUUUGAAUUUAUUUUUGUGACCACGAGAAUAUUGCACAAAAAAUAACCGCUUGUUU-GCCGUAAAUUUAGUC ...........-..(((((..(.(((--.........((((....(((((((((....))))))))).....))))((((((....))))))-))))..))))).... ( -15.50, z-score = -0.58, R) >droSec1.super_18 848825 104 - 1342155 CCAAUCGCCCCGCCAAAAUCACUGGC--ACUCACCUCUGUGAAUUUAUUUUUGUGACCACGAAAAUAUUGCACAAAAAGUUCCAGCUUGUUUGGCCGUAAAUUUGG-- ((((.......((((((....((((.--(((......((((....(((((((((....)))))))))...))))...))).))))....)))))).......))))-- ( -27.94, z-score = -3.24, R) >droSim1.chr2L 20530300 104 - 22036055 CCAAUCGCCCCGCCAAAAUCGCUGGC--ACUCACCUCUGUGAAUUUAUUUUUGUGACCACGAAAAUAUUGCACAAAAAGUGCCAGCUUGUUUGGCCGUAAAUUUGG-- ((((.......((((((...((((((--(((......((((....(((((((((....)))))))))...))))...)))))))))...)))))).......))))-- ( -38.24, z-score = -5.89, R) >consensus CCAAUCGCCCCGCCAAAAUCGCUGGC__ACUCACCUCUGUGAAUUUAUUUUUGUGACCACGAAAAUAUUGCACAAAAAAUGCCAGCUUGUUUGGCCGUAAAUUUGG__ ((((.....(.((((((...((((((....(((......)))...(((((((((....))))))))).............))))))...)))))).).....)))).. (-18.51 = -19.95 + 1.44)


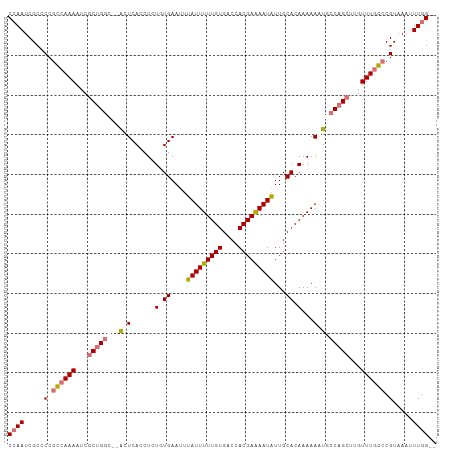
| Location | 20,915,049 – 20,915,157 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.97 |
| Shannon entropy | 0.14508 |
| G+C content | 0.43137 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.82 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20915049 108 - 23011544 GGUCCAAUCGCCCCGCCAAAAUCGCUGGCUUGCUCACCUCUUUGAAAUUAUUUUUGUGACCACGAAAAUGUUGCACAAAAAAUGCCAGCUUGUUUGGCCGUAAAUUUG (((......)))..((((((...((((((....(((......))).....((((((((.(.((......)).)))))))))..))))))...)))))).......... ( -29.00, z-score = -2.59, R) >droEre2.scaffold_4845 19037444 104 + 22589142 GGUCCAAUCGCCCC-UAAAAUUCGCUGGC--ACUCACCUCUUUGAAUUUAUUUUUGUGACCACGAGAAUAUUGCACAAAAAAUAACCGCUUGUUU-GCCGUAAAUUUA (((......)))..-..(((((..(.(((--.........((((....(((((((((....))))))))).....))))((((((....))))))-))))..))))). ( -16.80, z-score = -0.64, R) >droSec1.super_18 848826 106 - 1342155 GGUCCAAUCGCCCCGCCAAAAUCACUGGC--ACUCACCUCUGUGAAUUUAUUUUUGUGACCACGAAAAUAUUGCACAAAAAGUUCCAGCUUGUUUGGCCGUAAAUUUG (((......)))..((((((....((((.--(((......((((....(((((((((....)))))))))...))))...))).))))....)))))).......... ( -26.90, z-score = -2.66, R) >droSim1.chr2L 20530301 106 - 22036055 GGUCCAAUCGCCCCGCCAAAAUCGCUGGC--ACUCACCUCUGUGAAUUUAUUUUUGUGACCACGAAAAUAUUGCACAAAAAGUGCCAGCUUGUUUGGCCGUAAAUUUG (((......)))..((((((...((((((--(((......((((....(((((((((....)))))))))...))))...)))))))))...)))))).......... ( -37.20, z-score = -5.33, R) >consensus GGUCCAAUCGCCCCGCCAAAAUCGCUGGC__ACUCACCUCUGUGAAUUUAUUUUUGUGACCACGAAAAUAUUGCACAAAAAAUGCCAGCUUGUUUGGCCGUAAAUUUG (((......)))..((((((...((((((....(((......)))...(((((((((....))))))))).............))))))...)))))).......... (-18.64 = -19.82 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:27 2011