
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,852,754 – 20,852,856 |
| Length | 102 |
| Max. P | 0.716852 |

| Location | 20,852,754 – 20,852,856 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 60.26 |
| Shannon entropy | 0.85982 |
| G+C content | 0.50120 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -11.88 |
| Energy contribution | -11.69 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.37 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20852754 102 + 23011544 GCGGUGCCGAUGAAAGCCUUUCUGGCUGGUCUCUUCUUGCUGGCCAAUGCCUGGCACAUCACAGGUGAG----AUGUGUUAAACGUCGAAAAUAAUAAAGUGUCGA .(((..((((((..((((.....)))).(((((.(((((.((((((.....)))).)).)).))).)))----))........)))))...........)..))). ( -28.80, z-score = -0.61, R) >droSim1.chr2L 20459797 102 + 22036055 GCGGUGCCGAUGAAAGCCUUUCUGGCUGGUCUCUUCCUGCUGGCCAAUGCCUGGCACAUCACAGGUGAG----AUGUGUUAAACGUCCAAAACAACAAAGUGUCGA .(((..(.((((...(((....((((..((........))..))))......))).))))...(.((.(----((((.....)))))))...)......)..))). ( -28.60, z-score = 0.01, R) >droSec1.super_18 769382 102 + 1342155 GCGGUGCCGAUGAAAGCCUUUCUGGCUGGUCUCUUCCUGCUGGCCAAUGCCUGGCACAUCACAGGUGAG----AUGUGUUAAACGUCCAAAACAACAAAGUGUCGA .(((..(.((((...(((....((((..((........))..))))......))).))))...(.((.(----((((.....)))))))...)......)..))). ( -28.60, z-score = 0.01, R) >droYak2.chr2R 19247536 94 - 21139217 GCGGUGCCGAUGAAAGCCUUUCUGGCUGGCGUUUUCUUGCUGGCCAAUGCCUGGCACAUCACAGGUGAG----AUGUGUUCAA-------CGUC-CACAGUGUCGA .(((..(.((((...(((....((((..(((......)))..))))......))).))))....(((.(----((((.....)-------))))-))).)..))). ( -35.40, z-score = -1.36, R) >droEre2.scaffold_4845 18967525 93 - 22589142 GCGGUGCCGAUGAGAGCCUUUCUGGCUGGUGUCUUUUUGCUGGCCAAUGCCUGGCACAUCACAGGUGAG----CUGUGUUCCG-------CGUC-C-CAGUGUCGA ((((((((((((..((((.....))))..))))........(((....))).)))))..(((((.....----)))))...))-------)...-.-......... ( -31.70, z-score = 0.25, R) >droAna3.scaffold_12943 1165838 101 - 5039921 GCCAUGUCCAUGAAAGCCCUUUUGGCCACCACUUUCUUGCUGGCCAAUGCCUGGUCCAUCACAGGUGAG----AAAUGUUUCUGUUUUUUCACC-UCAAGUGUCGA ..(((....)))...(((...(((((((.((......)).))))))).....))).(..((((((((((----(((.........)))))))))-)...)))..). ( -26.00, z-score = -0.82, R) >droWil1.scaffold_180708 8848712 103 + 12563649 ---GUGCUGGUGAAAUUGAUCAUUGCUGGUGUUUUCCUUCUGGCCAAUGCCUUUCACAUUCAAGGUGAGUUCAAUCUGUGAGAAAUUUUCUAAUAAAAAGUGUAGA ---....(((.((((((..((((....((((((..((....))..)))))).(((((.......)))))........))))..))))))))).............. ( -16.80, z-score = 1.44, R) >droVir3.scaffold_12963 9413809 81 - 20206255 ---GCACCCAUGAAAUUCAUAAUUGCUGGCUUUUGCCUGUUGGCCAGCGCCUACAAUAUUAAAGGUGAGUUC-CG----CUCGUUCCGA----------------- ---((....(((.....)))....(((((((..........))))))))).............((((((...-..----))))..))..----------------- ( -18.60, z-score = -0.21, R) >droMoj3.scaffold_6500 12845434 102 - 32352404 ---GCGCCCAUUAAGUUAAUAAUUGCUGGCUUCUGCCUGCUGGCCACCGCCUACAAUAUCCGAGGUGAGUUC-CGAUUCCCCAUCUCGCCGAUCUGCGUUCUCCAU ---((((..(((.........((((..(((....(((....)))....)))..))))......((((((...-...........)))))))))..))))....... ( -24.14, z-score = -0.79, R) >droGri2.scaffold_15252 12213473 86 - 17193109 ---GCGCCUAUUAAAUUUAUUAUUGCUGGCUUUUGCCUGCUGGCUACCGCCUACAACAUCAGCGGUGAGUUAAUAGUCGAUCGUCACAA----------------- ---.((.(((((((.....(((((((((((....)))((..(((....)))..)).....)))))))).))))))).))..........----------------- ( -23.70, z-score = -1.61, R) >consensus GCGGUGCCGAUGAAAGCCUUUCUGGCUGGCCUCUUCCUGCUGGCCAAUGCCUGGCACAUCACAGGUGAG____AUGUGUUACACGUCGA_AAUC_CAAAGUGUCGA ........................((((((........))))))...((((((........))))))....................................... (-11.88 = -11.69 + -0.19)

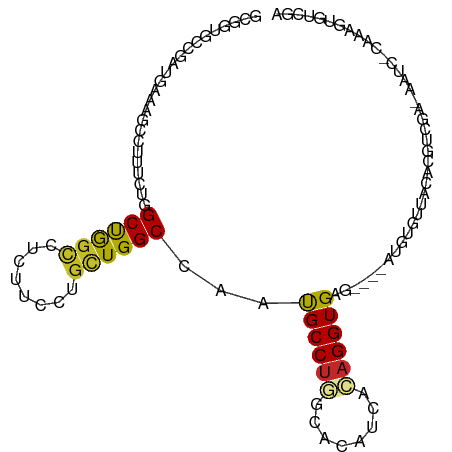
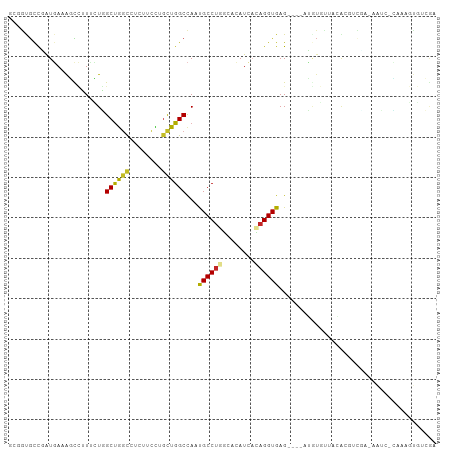
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:24 2011