| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,833,099 – 20,833,215 |
| Length | 116 |
| Max. P | 0.500000 |

| Location | 20,833,099 – 20,833,215 |
|---|---|
| Length | 116 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.55524 |
| G+C content | 0.54690 |
| Mean single sequence MFE | -40.69 |
| Consensus MFE | -20.36 |
| Energy contribution | -19.95 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.73 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 20833099 116 + 23011544 CAAGCAUCCUGCCCAGCAUU-AGUGAGUUUUCGGCCAUCUGCUCGGCACUCGAGGGCGUCAGGGCCUUCAGGUGCUGCAGGUAGCUCACCUUGUCGAUUUGCAGUGCUGCCGAGUAG ..........(((.(((...-.....)))...)))...(((((((((....((((((......)))))).((..((((((((.((.......))..))))))))..))))))))))) ( -44.70, z-score = -0.98, R) >droSim1.chr2L 20440795 116 + 22036055 CAAGCAUCCUGCCCAGCAUU-AGCGAGUUCUCGGCCAUCUGCUCGGCACUCGAGGGCGUCAGGGCCUUCAGGUGCUGCAGGUAGCUCACCUUGUCGAUCUGCAGUGCUGCCGAGUAG ..........(((..((...-.))((....)))))...(((((((((....((((((......)))))).((..((((((((.((.......))..))))))))..))))))))))) ( -49.30, z-score = -1.66, R) >droSec1.super_18 751156 116 + 1342155 CAAGCAUCCUGCCCAGCAUU-AGCGAGUUCUCGGCCAUCUGCUCGGCACUCGAGGGCGUGAGGGCCUUCAGGUGCUGCAGAUAGCUCACCUUGUCGAUUUGCAGUGCUGCCGAGUAG ..........(((..((...-.))((....)))))...(((((((((....((((((......)))))).((..((((((((.((.......))..))))))))..))))))))))) ( -47.30, z-score = -1.25, R) >droEre2.scaffold_4845 18950167 116 - 22589142 CAAGCAUCCUGCCCAGCAUU-AGUGAGUUCUCGGCCAUCUGCUCGGCACUCGAGGGCGUGAGGGUCUUCAGGUGCUGUAGGUAGCUCACCUUGUCGAUCUGCAGUGCUGCCGAGUAG ..........(((.((.(((-....))).)).)))...(((((((((....((((((......)))))).((..((((((((.((.......))..))))))))..))))))))))) ( -43.80, z-score = -0.58, R) >droYak2.chr2R 19224299 116 - 21139217 CAAGCAUCCUGCCCAGCAUU-AGUGAGUUCUCAGCCAUCUGCUCGGCACUUGAGGGAGUCAGGGCCUUCAGGUGUUGUAGGUAGCUCACCUUGUCGAUUUGCAGUGCUGCCGAGUAA ...((.....)).(((((((-.(..((((..((((.((((((...(((((((((((........))))))))))).)))))).))......))..))))..))))))))........ ( -38.40, z-score = -0.01, R) >droAna3.scaffold_12943 1148119 116 - 5039921 CCAACAUCCUCCCCAACAUC-AGAGAAUUCUCCGCCAUCUGCUCGGCACUCGAGGGCGUCAGUGUCUUCAGGUGCUGCAGGUAGCUGACCUUGUCGAUCUGCAGGGCGGCGGCGUAG ....................-.(((....)))((((((((((..(((((..(((((((....)))))))..))))))))))).((((.((((((......))))))))))))))... ( -46.90, z-score = -2.12, R) >dp4.chr4_group3 4238058 116 - 11692001 CCAACAUCCUUCCCAGCAUC-AAGGAGUUCUCGGCCAUCUGCUCGGCACUGGACGGGGUUAAUGUCUUUAGGUGCUGCAGAUAGCUGACCUUGUCGAUCUGCAGAGCAGCCGAGUAU ..(((.(((((.........-))))))))((((((....(((((.(((.(.((((((((((.(((((.(((...))).)))))..)))))))))).)..))).)))))))))))... ( -43.50, z-score = -1.84, R) >droPer1.super_1 1379406 116 - 10282868 CCAACAUCCUUCCCAGCAUC-AAGGAGUUCUCGGCCAUCUGCUCGGCACUGGACGGGGUCAAAGUCUUUAGGUGCUGCAGAUAGCUGACCUUGUCGAUCUGCAAAGCAGCCGAGUAU ..(((.(((((.........-))))))))((((((....((((..(((.(.((((((((((..((((.(((...))).))))...)))))))))).)..)))..))))))))))... ( -40.50, z-score = -1.39, R) >droVir3.scaffold_12963 9382098 116 - 20206255 CGAGCAGCCGGCCCAGCAUU-AGCGAGUUCUCGGCCAUCUGCUCGGCACUCGAGGGCGCCAGGGUCUUCAGGUGCUGUAGGUAGCUAACCUUGUCGAUCUGCAGGGCUGCCGAGUAG ((.((((((((((..((...-.))((....))))))..((((.((((((..((((((......))))))..)))))).((((.....)))).........))))))))))))..... ( -50.30, z-score = -1.08, R) >droGri2.scaffold_15252 12195058 116 - 17193109 CGAGCGUUCUGCCCAGCAUU-AGAGAAUUCUCGGCCAUUUGCUCAGCGCUCGAGGGCGACAAGGUCUUCAGGUGCUGCAGAUAGCUAACCUUGUCGAUCUGCAAAGCAGCCGAAUAG .(((((((..((...((...-.(((....))).)).....))..)))))))((((((......)))))).((((((((((((.((.......))..))))))..)))).))...... ( -39.40, z-score = -0.60, R) >droWil1.scaffold_180708 8821854 116 + 12563649 CCAACAUACGACCCAACAUC-AAAGAAUUUUCAGCCAUUUGUUCGGCACUCGAUGGCGUCAAAGUCUUCAGAUGUUGCAGAUAACUGACCUUAUCAAUUUGCAAAGCGGCUGAGUAG ....................-........(((((((.(((((.(((((((.((.(((......))).)))).)))))..(((((......))))).....)))))..)))))))... ( -24.70, z-score = -0.37, R) >droMoj3.scaffold_6500 12822448 116 - 32352404 CGAGCAAGCGGCCCAGCAUC-AGCGAGUUCUCGGCCAUCUGCUCGGCGCUCGAUGGUGACAACGUCUUUAGGUACUGCAGAUAGCUCACUUUGUCGAUCUGCAGGGCAGCCGAGUAG .........((((..((...-.))((....))))))..((((((((((((((.((....)).))..........((((((((.((.......))..))))))))))).))))))))) ( -45.00, z-score = -1.23, R) >anoGam1.chr3R 2200079 116 - 53272125 CUUGAAUGCGACCAUUCAUA-AUCGAGUUUUCGGCCAUAUGCUCGGGACUCGAAGCGGGCAGCGACUUGAUGUAGUUCAAAUAGUUCACUUUGUCUAUCUGCAACGAAGCGGAAAAU ..((((((....))))))..-.((((((((..(((.....)))..))))))))...((((((.(((((((......)))...))))....)))))).(((((......))))).... ( -29.80, z-score = -0.22, R) >triCas2.ChLG5 8987904 113 - 18847211 CUUGC-UGCGCCCCACCAAGGAGGGCAAUUGCGGCUUUUUGUUGAAUUUU---GUUCGUCAAACUCUUUGCAUAUUGGAUGUAAAACACUUUAUCAAAAUGAUUAAUAUUGGCGAAA ...((-((((((((......).))))....)))))...............---.((((((((....(((((((.....))))))).......(((.....))).....)))))))). ( -26.00, z-score = -0.47, R) >consensus CAAGCAUCCUGCCCAGCAUU_AGCGAGUUCUCGGCCAUCUGCUCGGCACUCGAGGGCGUCAGGGUCUUCAGGUGCUGCAGAUAGCUCACCUUGUCGAUCUGCAGAGCAGCCGAGUAG .............................((((((...((((.(((((((.((.((........)).)).)))))))..(((((......))))).....))))....))))))... (-20.36 = -19.95 + -0.41)

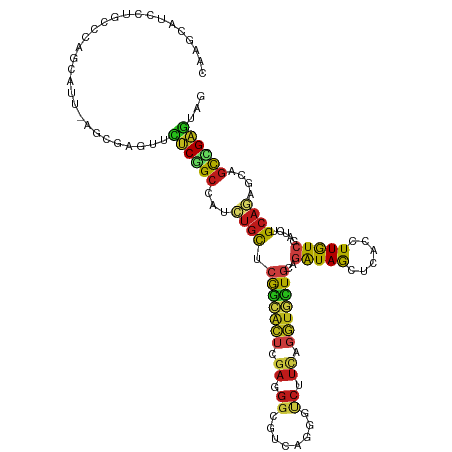

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:23 2011