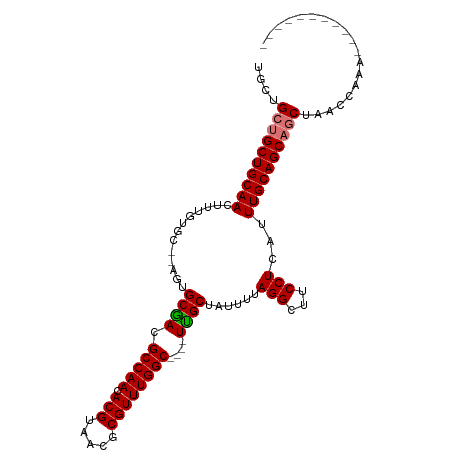
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,809,852 – 20,809,944 |
| Length | 92 |
| Max. P | 0.968289 |

| Location | 20,809,852 – 20,809,944 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.84 |
| Shannon entropy | 0.23046 |
| G+C content | 0.49602 |
| Mean single sequence MFE | -35.79 |
| Consensus MFE | -34.00 |
| Energy contribution | -33.14 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
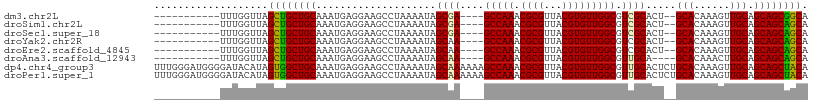
>dm3.chr2L 20809852 92 + 23011544 -----------UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCGA----GCCAAACGCGUUACGUGUUGGCGUCGCACU--GCACAAAGUUGCAGCAGCGGCA -----------....(((.((((((((((.(((((...)))......((((----(((((.((((...))))))))).))))...--...))...))))))))))))). ( -37.40, z-score = -1.98, R) >droSim1.chr2L 20416881 92 + 22036055 -----------UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCGA----GCCAAACGCGUUACGUGUUGGCGUCGCACU--GCACAAAGUUGCAGCAGCAGCA -----------....(((.((((((((((.(((((...)))......((((----(((((.((((...))))))))).))))...--...))...))))))))))))). ( -38.20, z-score = -2.49, R) >droSec1.super_18 720643 92 + 1342155 -----------UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCGA----GCCAAACGCGUUACGUGUUGGCGUCGCACU--GCACAAAGUUGCAGCAGCAGCA -----------....(((.((((((((((.(((((...)))......((((----(((((.((((...))))))))).))))...--...))...))))))))))))). ( -38.20, z-score = -2.49, R) >droYak2.chr2R 11387587 92 - 21139217 -----------UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCAA----GCCAAACGCGUUACGUGUUGGCGUCGCACU--GCACAAAGUUGCAGCAGCAGCA -----------....(((.((((((((((.(((((...)))......((..----(((((.((((...)))))))))...))...--...))...))))))))))))). ( -33.90, z-score = -1.24, R) >droEre2.scaffold_4845 18929377 92 - 22589142 -----------UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCAA----GCCAAACGCGUUACGUGUUGGCGUCGCACU--GCACAAAGUUGCAGCAGCAGCA -----------....(((.((((((((((.(((((...)))......((..----(((((.((((...)))))))))...))...--...))...))))))))))))). ( -33.90, z-score = -1.24, R) >droAna3.scaffold_12943 1125544 90 - 5039921 -----------UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCAA----GCCAAACGCGUUACGUGUUGGCGUUGCA----GCACAAACUUGCAGCAGCAGCA -----------....(((.((((((((((...(((...)))......((((----(((((.((((...))))))))).)))).----........))))))))))))). ( -36.10, z-score = -2.35, R) >dp4.chr4_group3 4219086 109 - 11692001 UUUGGGAUGGGGAUACAUAGUGGCUGCAAAUGAGGAAGCCUAAAAUAGCAAAAAAGCCAAACGCGUUACGUGUUGGCGUUGCACUCUGCACAAAGUUGCAGCAGCUACA ......(((......))).((((((((....................((((....(((((.((((...))))))))).))))....((((......)))))))))))). ( -34.30, z-score = -1.23, R) >droPer1.super_1 1360283 109 - 10282868 UUUGGGAUGGGGAUACAUAGUGGCUGCAAAUGAGGAAGCCUAAAAUAGCAAAAAAGCCAAACGCGUUACGUGUUGGCGUUGCACUCUGCACAAAGUUGCAGCAGCUACA ......(((......))).((((((((....................((((....(((((.((((...))))))))).))))....((((......)))))))))))). ( -34.30, z-score = -1.23, R) >consensus ___________UUUGGUUAGCUGCUGCAAAUGAGGAAGCCUAAAAUAGCAA____GCCAAACGCGUUACGUGUUGGCGUCGCACU__GCACAAAGUUGCAGCAGCAGCA ...................((((((((....................((((....(((((.((((...))))))))).)))).....(((......))).)))))))). (-34.00 = -33.14 + -0.86)
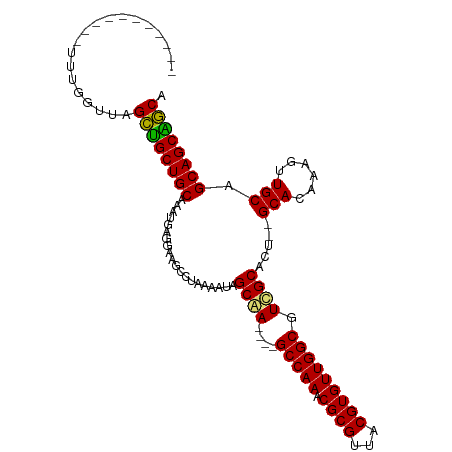
| Location | 20,809,852 – 20,809,944 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.84 |
| Shannon entropy | 0.23046 |
| G+C content | 0.49602 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -28.82 |
| Energy contribution | -28.85 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20809852 92 - 23011544 UGCCGCUGCUGCAACUUUGUGC--AGUGCGACGCCAACACGUAACGCGUUUGGC----UCGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA----------- ....((((((((((........--...((((.(((((.(((.....))))))))----))))......(((...)))...))))))))))........----------- ( -33.90, z-score = -2.28, R) >droSim1.chr2L 20416881 92 - 22036055 UGCUGCUGCUGCAACUUUGUGC--AGUGCGACGCCAACACGUAACGCGUUUGGC----UCGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA----------- .((((((((((((......)))--)..((((.(((((.(((.....))))))))----))))....................))))))))........----------- ( -35.00, z-score = -2.58, R) >droSec1.super_18 720643 92 - 1342155 UGCUGCUGCUGCAACUUUGUGC--AGUGCGACGCCAACACGUAACGCGUUUGGC----UCGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA----------- .((((((((((((......)))--)..((((.(((((.(((.....))))))))----))))....................))))))))........----------- ( -35.00, z-score = -2.58, R) >droYak2.chr2R 11387587 92 + 21139217 UGCUGCUGCUGCAACUUUGUGC--AGUGCGACGCCAACACGUAACGCGUUUGGC----UUGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA----------- .((((((((((((......)))--)..((((.(((((.(((.....))))))))----))))....................))))))))........----------- ( -32.90, z-score = -1.78, R) >droEre2.scaffold_4845 18929377 92 + 22589142 UGCUGCUGCUGCAACUUUGUGC--AGUGCGACGCCAACACGUAACGCGUUUGGC----UUGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA----------- .((((((((((((......)))--)..((((.(((((.(((.....))))))))----))))....................))))))))........----------- ( -32.90, z-score = -1.78, R) >droAna3.scaffold_12943 1125544 90 + 5039921 UGCUGCUGCUGCAAGUUUGUGC----UGCAACGCCAACACGUAACGCGUUUGGC----UUGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA----------- ....((((((((((((..(.((----(((((.(((((.(((.....))))))))----)))).......)))...)..))))))))))))........----------- ( -34.71, z-score = -2.79, R) >dp4.chr4_group3 4219086 109 + 11692001 UGUAGCUGCUGCAACUUUGUGCAGAGUGCAACGCCAACACGUAACGCGUUUGGCUUUUUUGCUAUUUUAGGCUUCCUCAUUUGCAGCCACUAUGUAUCCCCAUCCCAAA .((((..(((((((......((.((((((((.(((((.(((.....))))))))....)))).))))...))........)))))))..))))................ ( -27.24, z-score = -0.73, R) >droPer1.super_1 1360283 109 + 10282868 UGUAGCUGCUGCAACUUUGUGCAGAGUGCAACGCCAACACGUAACGCGUUUGGCUUUUUUGCUAUUUUAGGCUUCCUCAUUUGCAGCCACUAUGUAUCCCCAUCCCAAA .((((..(((((((......((.((((((((.(((((.(((.....))))))))....)))).))))...))........)))))))..))))................ ( -27.24, z-score = -0.73, R) >consensus UGCUGCUGCUGCAACUUUGUGC__AGUGCGACGCCAACACGUAACGCGUUUGGC____UUGCUAUUUUAGGCUUCCUCAUUUGCAGCAGCUAACCAAA___________ ....((((((((((.............((((.(((((.(((.....))))))))....))))......(((...)))...))))))))))................... (-28.82 = -28.85 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:22 2011