| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,789,058 – 20,789,162 |
| Length | 104 |
| Max. P | 0.808842 |

| Location | 20,789,058 – 20,789,162 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 58.18 |
| Shannon entropy | 0.82186 |
| G+C content | 0.33522 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -7.04 |
| Energy contribution | -6.94 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.92 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
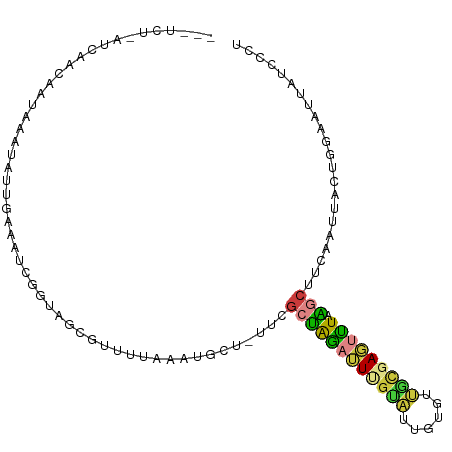
>dm3.chr2L 20789058 104 + 23011544 ACGUCUAAUCAACAAUAAAUAUUGAAAUCGCUAGCGUUUUAAAUGCUCUUCGCUAAAUUUGUAUUGUGUUGCGAGUUUAAGCUUCAAUUACUGGAAUUAUCCCU ....................((((((......(((((.....)))))....((((((((((((......))))))))).)))))))))....(((....))).. ( -17.20, z-score = -0.75, R) >droSim1.chr2L 20392400 102 + 22036055 --AUCUAAUCAACAAUAAAUAUUGAAAUCGGUAGCGUUUUAAAUGCUAUUCGCUAGAUUUGCAUUGUGUUGCGAGUUUAAGCUUCAAUUACUGGAAUUAUCCCU --..................((((((...((((((((.....)))))))).((((((((((((......))))))))).)))))))))....(((....))).. ( -22.60, z-score = -2.00, R) >droSec1.super_18 699712 102 + 1342155 --AUCUAAACAACAAUAAAUAUGGAAAUCGGUAGCGUUUUAAAUGCUAUUCGCUAGAUUUGCAUUGUGUUGCGAGUUUAAGCUUCAAUUACUGGCAUUAUCCCU --....................(((....((((((((.....)))))))).((((((((((((......))))))))).))).................))).. ( -20.90, z-score = -0.86, R) >droYak2.chr2R 11365694 99 - 21139217 --AUCUUAUCAACAAUCAAUAUGGAAAUC--UAGCGUUUCUAUUGCU-UUCGCUAGAUUUGUAUUGUGCUGCGAGUCUAAGCUUCAAUUACUGGAAUUAUCCCU --..............((((((..(((((--(((((...........-..))))))))))))))))((..((........))..))......(((....))).. ( -18.52, z-score = -0.84, R) >droEre2.scaffold_4845 18905061 93 - 22589142 ----------AACAAUAAAUAUUGAAAUCUGCAGCGUUUCUUAUGCUUUUCGCUAGAUUCGUAUUGUGCUGCGAGUUUAGGCUUCAAUUACUGGAAUUAUCCC- ----------..........((((((....(((((((.....))))).....(((((((((((......)))))))))))))))))))....(((....))).- ( -20.20, z-score = -1.25, R) >droMoj3.scaffold_6500 21306543 81 + 32352404 --------GCGGAGGGCAUUUUAAAAUUAUACAAUAUUGCAUACACA------UAGAGUUAUGUUAUAUAUAUAUGUUGGAAUUCGAUAGACACU--------- --------((.....))(((((((...((((..((((.(((((.((.------....))))))).)))).))))..)))))))............--------- ( -8.90, z-score = 0.60, R) >droVir3.scaffold_12963 16378001 80 + 20206255 ---------------GGAGCAUACAAAUUAAAAACAGCUUGCAAAUG----GACGAAUUCGUUUCAUACAAUAACAUUAGACUUAAGCUUAAGGAAACC----- ---------------.((((................(....)..(((----((((....)).)))))...................))))..(....).----- ( -8.00, z-score = -0.11, R) >consensus ___UCU_AUCAACAAUAAAUAUUGAAAUCGGUAGCGUUUUAAAUGCU_UUCGCUAGAUUUGUAUUGUGUUGCGAGUUUAAGCUUCAAUUACUGGAAUUAUCCCU ...................................................((((((((((((......))))))))).)))...................... ( -7.04 = -6.94 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:17 2011