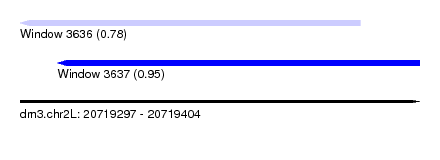
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,719,297 – 20,719,404 |
| Length | 107 |
| Max. P | 0.946384 |

| Location | 20,719,297 – 20,719,388 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 72.73 |
| Shannon entropy | 0.42741 |
| G+C content | 0.49929 |
| Mean single sequence MFE | -17.23 |
| Consensus MFE | -14.00 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
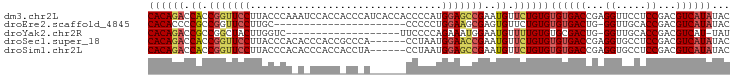
>dm3.chr2L 20719297 91 - 23011544 CUUACCCAAAUCCACCACCCAUCACCACCCCAUGGAGCCGAAUGUUCUGUGUGUGACCGAGGUUCCUCCGACGUCAUAUACUCGCUUUAUA ...............................(((((((.((....)).(((((((((.(((....)))....)))))))))..))))))). ( -18.40, z-score = -1.26, R) >droYak2.chr2R 11302198 70 + 21139217 ------------CUUGGU------CUUCCCCA-GAAAUGGAAUGUUUUGUGUGCGACUG-GGUUGCACCGACGUCAU-UAUACCCUUUAAA ------------...(((------.....(((-....)))((((..(((.(((((((..-.))))))))))...)))-)..)))....... ( -17.30, z-score = -1.06, R) >droSec1.super_18 638194 85 - 1342155 CUUACCCACACCCACCGC------CCACCUAAUGGAACCGAAUGUUCUGUGUGUGACCGAGGUGCCUCCGACGUCAUAUACUCACUUUAUA ..................------.........(((((.....)))))(((((((((.(((....)))....))))))))).......... ( -16.60, z-score = -0.76, R) >droSim1.chr2L 20332779 85 - 22036055 CUUACCCACACCCACCAC------CUACCUAAUGGAGCCGAAUGUUCUGUGUGUGACCGAGGUGCCUCCGACGUCAUAUACUCACUUUAUA ..................------.........(((((.....)))))(((((((((.(((....)))....))))))))).......... ( -16.60, z-score = -0.62, R) >consensus CUUACCCACACCCACCAC______CCACCCAAUGGAACCGAAUGUUCUGUGUGUGACCGAGGUGCCUCCGACGUCAUAUACUCACUUUAUA .................................(((((.....)))))(((((((((...((.....))...))))))))).......... (-14.00 = -13.50 + -0.50)
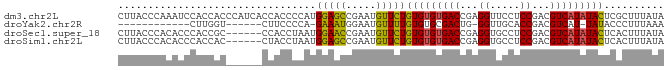
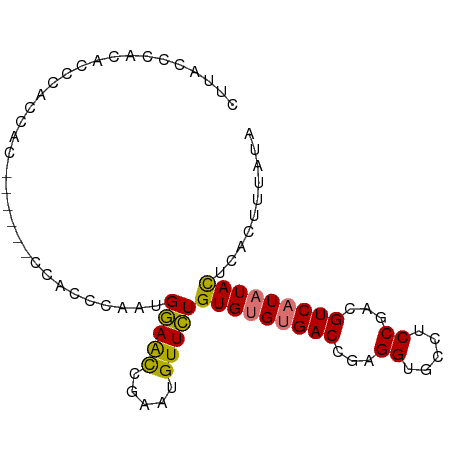
| Location | 20,719,307 – 20,719,404 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 71.91 |
| Shannon entropy | 0.46422 |
| G+C content | 0.56460 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
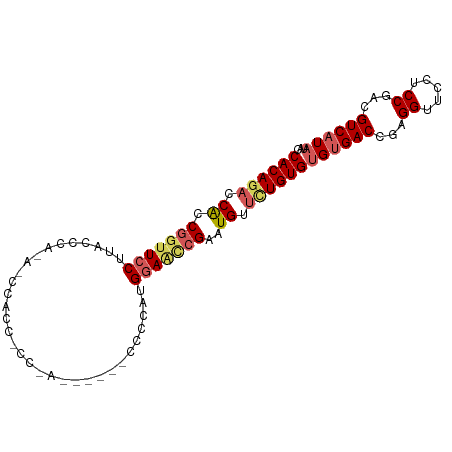
>dm3.chr2L 20719307 97 - 23011544 CACAGACCACCGGUUCCUUACCCAAAUCCACCACCCAUCACCACCCCAUGGAGCCGAAUGUUCUGUGUGUGACCGAGGUUCCUCCGACGUCAUAUAC ((((((.((.(((((((................................)))))))..)).))))))((((((.(((....)))....))))))... ( -26.05, z-score = -2.21, R) >droEre2.scaffold_4845 18844091 74 + 22589142 CACACCCCGCCGGUUCCUUGC----------------------CCCCCUGGAAGCGAGUGUUCUGUGUGUGACUG-GGUUGCACCGACGUCAUAUAC .......(((((.((((....----------------------......)))).)).)))....((((((((((.-((.....)).).))))))))) ( -22.40, z-score = -0.52, R) >droYak2.chr2R 11302208 76 + 21139217 CACAGACCGCCGGCUACUUGGUC-------------------UUCCCCAGAAAUGGAAUGUUUUGUGUGCGACUG-GGUUGCACCGACGUCAU-UAU ...((((((.........)))))-------------------)...(((....)))((((..(((.(((((((..-.))))))))))...)))-).. ( -20.90, z-score = -0.31, R) >droSec1.super_18 638204 91 - 1342155 CACAGACCACCGGUUCCUUACCCACACCCACCGCCCA------CCUAAUGGAACCGAAUGUUCUGUGUGUGACCGAGGUGCCUCCGACGUCAUAUAC ((((((.((.(((((((....................------......)))))))..)).))))))((((((.(((....)))....))))))... ( -26.27, z-score = -2.16, R) >droSim1.chr2L 20332789 91 - 22036055 CACAGACCACCGGUUCCUUACCCACACCCACCACCUA------CCUAAUGGAGCCGAAUGUUCUGUGUGUGACCGAGGUGCCUCCGACGUCAUAUAC ((((((.((.(((((((....................------......)))))))..)).))))))((((((.(((....)))....))))))... ( -26.27, z-score = -2.14, R) >consensus CACAGACCACCGGUUCCUUACCCA_A_CCACC_CC_A______CCCCAUGGAACCGAAUGUUCUGUGUGUGACCGAGGUUCCUCCGACGUCAUAUAC ((((((.((.(((((((................................)))))))..)).))))))((((((...((.....))...))))))... (-17.45 = -18.41 + 0.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:13 2011