| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,714,518 – 20,714,608 |
| Length | 90 |
| Max. P | 0.966523 |

| Location | 20,714,518 – 20,714,608 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 65.90 |
| Shannon entropy | 0.66492 |
| G+C content | 0.45731 |
| Mean single sequence MFE | -14.91 |
| Consensus MFE | -8.00 |
| Energy contribution | -8.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
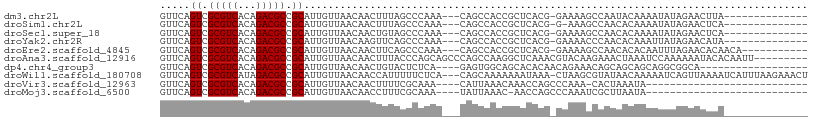
>dm3.chr2L 20714518 90 - 23011544 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUUUAGCCCAAA---CAGCCACCGCUCACG-GAAAAGCCAAUACAAAAUAUAGAACUUA-------------- ((((.((.(((((...))))).)).((((......((((.((.....---..))..(((....))-).)))).....))))......))))...-------------- ( -16.80, z-score = -1.80, R) >droSim1.chr2L 20328076 89 - 22036055 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUUUAGCCCAAA---CAGCCACCGCUCACG-G-AAAGCCAACACAAAAUAUAGAACUCA-------------- ((((.((.(((((...))))).)).((((......((((.((.....---..))..(((....))-)-)))).....))))......))))...-------------- ( -16.20, z-score = -1.66, R) >droSec1.super_18 633532 90 - 1342155 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUGUAGCCCAAA---CAGCCACCGCUCACG-GAAAAGCCAACACAAAAUAUAGAACUCA-------------- ((((.((.(((((...))))).))..((((.....((((.......)---)))...(((....))-).......)))).........))))...-------------- ( -18.00, z-score = -1.65, R) >droYak2.chr2R 11297456 90 + 21139217 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAAGUUCAGCCCAAA---CAGCCACCGCUCACG-GAAAACCCAACACAAAUUAUAGAACAUA-------------- ((((.((.(((((...))))).))..((((..........((.....---..))..(((....))-).......)))).........))))...-------------- ( -16.10, z-score = -2.07, R) >droEre2.scaffold_4845 18839705 93 + 22589142 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUUCAGCCCAAA---CAGCCACCGCUCACG-GAAAAGCCAACACACAAUUUAGAACACAACA----------- ((((.((.(((((...))))).))(((((...........((.....---..))..(((....))-)............)))))...))))......----------- ( -16.30, z-score = -1.69, R) >droAna3.scaffold_12916 7219830 99 - 16180835 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUUUACCCAGCAGCCCAGCCAAGGCUCAAACGUACAAGAAACUAAAUCCAAAAAAUACACAAUU--------- .....((.(((((...))))).))...........((((((.....((((.......)))).....))).)))..........................--------- ( -13.90, z-score = -1.01, R) >dp4.chr4_group3 4112324 86 + 11692001 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUGUACUCUCA----GAGUGGCAGCACACAACAGAAACAGCAGCAGCAGGCGGCA------------------ .....(((........)))(((((.(((((.....((((((((...----)))).)))).....))))).....((....))..))))).------------------ ( -24.00, z-score = -0.75, R) >droWil1.scaffold_180708 4445746 104 - 12563649 GUUCAGUCGCGUCAUAGACGCCGCAUUGUUAACAACCAUUUUUCUCA---CAGCAAAAAAAUAAA-CUAAGCGUAUAACAAAAAUCAGUUAAAAUCAUUUAAGAAACU .(((.((.(((((...))))).))....(((((....((((((....---..((...........-....)).......))))))..)))))..........)))... ( -10.98, z-score = -0.57, R) >droVir3.scaffold_12963 7723858 76 + 20206255 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUUUUCGCAAA----CAUUAAACAAACCAGCCCAAA-CACUAAAUA--------------------------- (((..((.(((((...))))).)).((((.((......)).)))).----.....))).............-.........--------------------------- ( -8.20, z-score = -0.45, R) >droMoj3.scaffold_6500 29450998 76 - 32352404 GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACCUUUCGCAAA----UAUUAAAC-AACCAGCCCAAAUCGCUUAAUA--------------------------- (((..((.(((((...))))).)).((((.((......)).)))).----.....)))-....(((.......))).....--------------------------- ( -8.60, z-score = -0.31, R) >consensus GUUCAGUCGCGUCACAGACGCCGCAUUGUUAACAACUUUAGCCCAAA___CAGCCACCGCUCACG_GAAAAGCCAACACAAAAUAUAGAACACA______________ .....((.(((((...))))).)).................................................................................... ( -8.00 = -8.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:11 2011