| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,713,120 – 20,713,229 |
| Length | 109 |
| Max. P | 0.746650 |

| Location | 20,713,120 – 20,713,229 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.65 |
| Shannon entropy | 0.45356 |
| G+C content | 0.58702 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.46 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
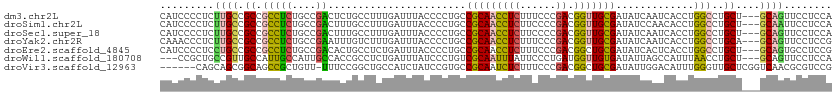
>dm3.chr2L 20713120 109 - 23011544 CAUCCCCUCUUGCCGCCGCCUCUGCCGACUCUGCCUUUGAUUUACCCCUGCCGCAACCUCUUUCCCGACGGUUGCGAUAUCAAUCACCUGGCCUGCU---GCAGUUCCUCCA .........((((.((.((....)).......(((..(((((.........(((((((((......)).))))))).....)))))...)))..)).---))))........ ( -22.34, z-score = -1.78, R) >droSim1.chr2L 20326707 109 - 22036055 CAUCCCCUCUUGCCGCCGCCUCUGCCGACUUUGCCUUUGAUUUACCCCUGCCGCAACCUCUUCCCCGACGGUUGCGAUAUCCAACACCUGGCCUGCU---GCAAUUCCUCCA .........((((.((.(((...((.......)).................(((((((((......)).))))))).............)))..)).---))))........ ( -22.20, z-score = -2.73, R) >droSec1.super_18 632172 109 - 1342155 CAUCCCCUCUUGCCGCCGCCUCUGCCGACUUUGCCUUUGAUUUACCCCUGCCGCAACCUCUUCCCCGACGGUUGCGAUAUCAAUCACCUGGCCUGCU---GCAGUUCCUCCA .........((((.((.((....)).......(((..(((((.........(((((((((......)).))))))).....)))))...)))..)).---))))........ ( -22.34, z-score = -1.82, R) >droYak2.chr2R 11296079 109 + 21139217 CAAACCCUCUUGCCGCCGCCUCUGCCGAAUUUGUCUUUGAUUUACCCCUGCCGCAACCUCUUUCCCGACGGUUGCGAUAUCAAUCACCUGGCCUGCA---GCAGUUCCUCCG .........((((.((.(((......(((((.......)))))........(((((((((......)).))))))).............)))..)).---))))........ ( -22.80, z-score = -1.60, R) >droEre2.scaffold_4845 18838420 109 + 22589142 CAUCCCCUCCUGCCGCCGCCUCUGCCGACACUGCCUCUGAUUUACCCCUGCCGCAACCUCUUUCCCGACGGCUGCGAUAUCACUCACCUGGCCUGCU---GCAGUGCCUCCG .........((((.((.(((...((.......)).................((((.((((......)).)).)))).............)))..)).---))))........ ( -19.90, z-score = -0.42, R) >droWil1.scaffold_180708 4444393 106 - 12563649 ---CCGCUGCCGUUGCCAUUGCCAUUGCCACCGCCUCUGAUUUAUCCCUGUCGCAAUUUAUUCCCUGAUGGUUGUGAUAUUAGCCAUUUAACCUGCU---GCAGUUCCUCCA ---..(((((.((....(((((....((....))....(((........)))))))).........((((((((......))))))))......)).---)))))....... ( -18.80, z-score = -0.45, R) >droVir3.scaffold_12963 7722486 105 + 20206255 ------CAGCAGCGGCAGCCGCUGUU-UUUCCGGCUGCCAUCUAUCCGUGCCGCAAUCUCUUUCCCGACGGCUGCGAUAUUGGACAUUUGGGUUGCUCGGUCAACGCGUCCG ------.((((((((((((((.....-....)))))))).....((((((.((((..(((......)).)..)))).)).)))).......))))))(((.(.....).))) ( -36.70, z-score = -0.82, R) >consensus CAUCCCCUCUUGCCGCCGCCUCUGCCGACUCUGCCUUUGAUUUACCCCUGCCGCAACCUCUUUCCCGACGGUUGCGAUAUCAAUCACCUGGCCUGCU___GCAGUUCCUCCA ...........((((..((....))..........................(((((((((......)).)))))))............))))((((....))))........ (-14.12 = -13.46 + -0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:11 2011