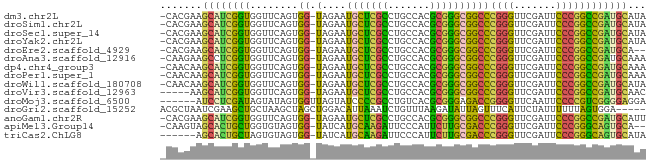
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,938,086 – 1,938,165 |
| Length | 79 |
| Max. P | 0.994438 |

| Location | 1,938,086 – 1,938,165 |
|---|---|
| Length | 79 |
| Sequences | 15 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 77.35 |
| Shannon entropy | 0.54456 |
| G+C content | 0.60506 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -24.93 |
| Energy contribution | -23.66 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
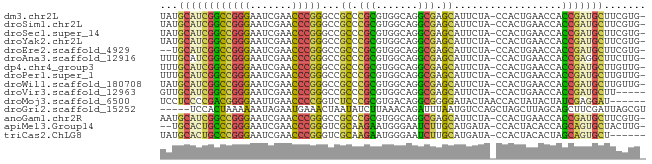
>dm3.chr2L 1938086 79 + 23011544 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUCGUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.75, R) >droSim1.chr2L 1905551 79 + 22036055 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUCGUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.75, R) >droSec1.super_14 1881386 79 + 2068291 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUCGUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.75, R) >droYak2.chr2L 1918421 79 + 22324452 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUCGUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.75, R) >droEre2.scaffold_4929 1983640 77 + 26641161 --UGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUCGUG- --.((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.73, R) >droAna3.scaffold_12916 532401 79 - 16180835 UUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAGGCUUCUUG- ...((.(((((((((.......)))))((((((.((...)).)))).))......-...........)))).))......- ( -27.90, z-score = -0.33, R) >dp4.chr4_group3 8696895 79 + 11692001 UUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUGUUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.91, R) >droPer1.super_1 10149415 79 + 10282868 UUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUGUUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.91, R) >droWil1.scaffold_180708 868112 79 - 12563649 UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUGUUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.93, R) >droVir3.scaffold_12963 5878357 75 + 20206255 GUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUU----- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))..----- ( -32.20, z-score = -2.00, R) >droMoj3.scaffold_6500 7541766 75 - 32352404 UCCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAU------ (((((.....(((((.......)))))(((.(((((.......))))))))..................))))).------ ( -28.10, z-score = -1.43, R) >droGri2.scaffold_15252 85780 76 + 17193109 -----UCCACUAAAAAAUAGAAUGAAACUAAUAUCUUAAACAGAUUUAAUGUCCAGCUAGCUUAGCAGCUUCGAUUAGCGU -----....((((....(((...((.......((((.....))))......))...)))..))))..(((......))).. ( -6.82, z-score = 1.09, R) >anoGam1.chr2R 5387131 79 - 62725911 AAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA-CCACUGAACCACCGAUGCUUCGUG- ...((((((((((((.......)))))((((((.((...)).)))).))......-...........)))))))......- ( -32.20, z-score = -1.67, R) >apiMel3.Group14 6137507 77 - 8318479 --UGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAUA-CCACUACACCAGCAGUGCUACUUG- --.((((((((((((.......)))))...((((((.......))))))......-...........)))))))......- ( -29.30, z-score = -3.54, R) >triCas2.ChLG8 3490830 74 - 15773733 UAUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAUA-CCACUACACUAGCAGUGCU------ ...((((((((((((.......)))))...((((((.......))))))......-...........))))))).------ ( -29.30, z-score = -3.84, R) >consensus UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUA_CCACUGAACCACCGAUGCUUCGUG_ ...((((((((((((.......)))))...((.(((.......))).))..................)))))))....... (-24.93 = -23.66 + -1.27)
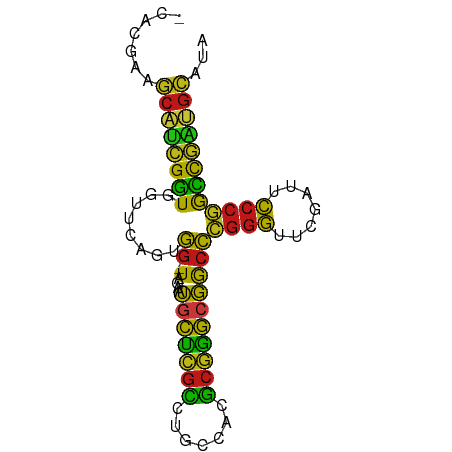
| Location | 1,938,086 – 1,938,165 |
|---|---|
| Length | 79 |
| Sequences | 15 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Shannon entropy | 0.54456 |
| G+C content | 0.60506 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -29.87 |
| Energy contribution | -28.04 |
| Covariance contribution | -1.83 |
| Combinations/Pair | 1.95 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
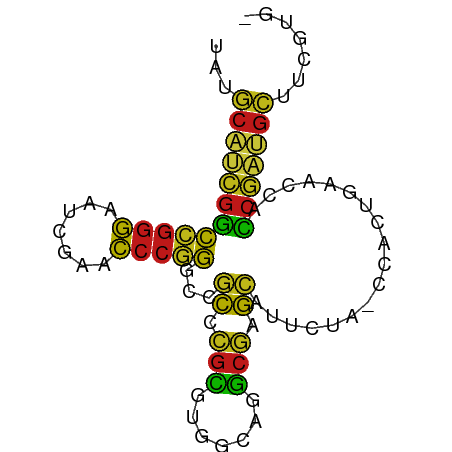
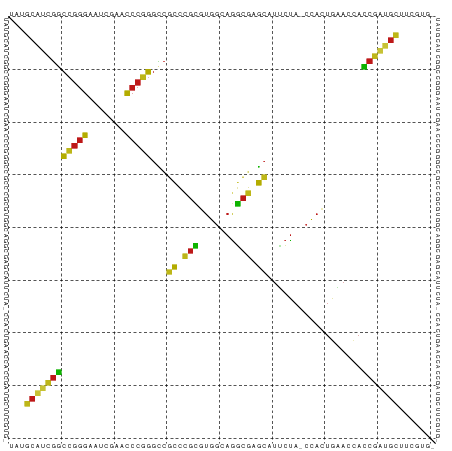
>dm3.chr2L 1938086 79 - 23011544 -CACGAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.63, R) >droSim1.chr2L 1905551 79 - 22036055 -CACGAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.63, R) >droSec1.super_14 1881386 79 - 2068291 -CACGAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.63, R) >droYak2.chr2L 1918421 79 - 22324452 -CACGAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.63, R) >droEre2.scaffold_4929 1983640 77 - 26641161 -CACGAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCA-- -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......)))))))))))).-- ( -35.90, z-score = -1.58, R) >droAna3.scaffold_12916 532401 79 + 16180835 -CAAGAAGCCUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA -...(((.((.....))))).((((-(((.........)))))))(((..(((((.(((.......)))))))).)))... ( -32.30, z-score = -0.59, R) >dp4.chr4_group3 8696895 79 - 11692001 -CAACAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.98, R) >droPer1.super_1 10149415 79 - 10282868 -CAACAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.98, R) >droWil1.scaffold_180708 868112 79 + 12563649 -CAACAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.96, R) >droVir3.scaffold_12963 5878357 75 - 20206255 -----AAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAC -----..(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -2.13, R) >droMoj3.scaffold_6500 7541766 75 + 32352404 ------AUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGGA ------.(((((((((.((.......)).))))(((.((((.....)))).(((.((((.......))))))))))))))) ( -30.80, z-score = -1.38, R) >droGri2.scaffold_15252 85780 76 - 17193109 ACGCUAAUCGAAGCUGCUAAGCUAGCUGGACAUUAAAUCUGUUUAAGAUAUUAGUUUCAUUCUAUUUUUUAGUGGA----- .((((((....((((((...)).)))).((.(((((((((.....)))).))))).))..........))))))..----- ( -14.90, z-score = -0.50, R) >anoGam1.chr2R 5387131 79 + 62725911 -CACGAAGCAUCGGUGGUUCAGUGG-UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUU -......(((((((..((((.....-..))))((.(((((((...)))))))))(((((.......))))))))))))... ( -35.90, z-score = -1.67, R) >apiMel3.Group14 6137507 77 + 8318479 -CAAGUAGCACUGCUGGUGUAGUGG-UAUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCA-- -......((((((((((((......-)))))(((((((.......)))))))..(((((.......)))))))))))).-- ( -32.20, z-score = -3.25, R) >triCas2.ChLG8 3490830 74 + 15773733 ------AGCACUGCUAGUGUAGUGG-UAUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAUA ------.(((((((...........-.....(((((((.......)))))))..(((((.......))))))))))))... ( -31.40, z-score = -3.83, R) >consensus _CACGAAGCAUCGGUGGUUCAGUGG_UAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA .......(((((((.................(((((((.......)))))))..(((((.......))))))))))))... (-29.87 = -28.04 + -1.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:41 2011