| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,712,876 – 20,712,972 |
| Length | 96 |
| Max. P | 0.895495 |

| Location | 20,712,876 – 20,712,972 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 61.85 |
| Shannon entropy | 0.68281 |
| G+C content | 0.46453 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -7.01 |
| Energy contribution | -7.09 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
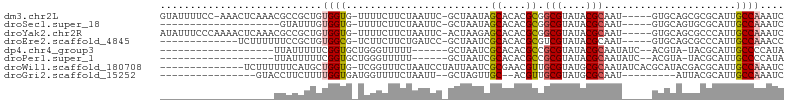
>dm3.chr2L 20712876 96 - 23011544 GUAUUUUCC-AAACUCAAACGCCGCUGUGGUG-UUUUCUUCUAAUUC-GCUAAUAGCACACGCGGCGUAUACGCAAU-----GUGCAGCGCGCAUUGCCAAAUC .........-........(((((((((((.((-((..(.........-)..)))).)))).)))))))....(((((-----((((...)))))))))...... ( -32.80, z-score = -3.46, R) >droSec1.super_18 631960 77 - 1342155 --------------------GUAUUUGUGGUG-UUUUCUUCUAAUUC-GCUAAUAGCACACGCGGCGUAUACGCAAU-----GUGCAGUGCGCAUUGCCAAAUC --------------------((((.(((.(((-(...((..((....-..))..))...)))).))).))))(((((-----((((...)))))))))...... ( -21.90, z-score = -0.99, R) >droYak2.chr2R 11295848 97 + 21139217 AUAUUUCCCAAAACUCAAACGCCGCUGUGGUG-UUUUCUUCUAAUUC-ACUAAGAGCACACGCGGCGUAUACGCAAU-----GUGCAGCGCCCAUUGCCAAAUC ..................(((((((.(((.((-((((..........-....))))))))))))))))....(((((-----(.((...)).))))))...... ( -27.24, z-score = -2.64, R) >droEre2.scaffold_4845 18838189 84 + 22589142 -------------UCUUUUUUCCGCUGUGGCG-UCUUCUUCUGAUCC-GCUAAUCGCACACGCGUCGUAUACGCAAU-----GUGCAGCGCCCAUUGCCAAACC -------------.........((((((((.(-((.......)))))-)).....(((((.((((.....))))..)-----)))))))).............. ( -21.60, z-score = -1.49, R) >dp4.chr4_group3 4110586 76 + 11692001 -------------------UUAUUUUUCGGUGCUGGGUUUUU------GCUAAUCGCACACGCCGCGUAUACGCAAUAUC--ACGUA-UACGCAUUGCCCCAUA -------------------.........((.((..(((.(.(------((.....))).).)))(((((((((.......--.))))-)))))...)).))... ( -24.00, z-score = -2.54, R) >droPer1.super_1 1252057 76 + 10282868 -------------------UUAUUUUUCGGUGCUGGGUUUUU------GCUAAUCGCACACGCCGCGUAUACGCAAUAUC--ACGUA-UACGCAUUGCCCCAUA -------------------.........((.((..(((.(.(------((.....))).).)))(((((((((.......--.))))-)))))...)).))... ( -24.00, z-score = -2.54, R) >droWil1.scaffold_180708 4444164 89 - 12563649 --------------UCUUUUUUCAUGCUGGUG-UCGGUUUCUAAUCCUAUUAAUCGCGAACGUUGCGUAUGCGCAAUAUCACGCAUACGACGCAUUGCCAAAUC --------------...........((..(((-((((........))........((((...))))(((((((........))))))))))))...))...... ( -21.80, z-score = -1.22, R) >droGri2.scaffold_15252 297483 75 - 17193109 ----------------GUACCUUCUUUUGGUGAUGGUUUUCUAAUU--GCUAGUUGC--ACGUUGCGUAUGCGCAAU---------AUUACGCAUUGCCAAAUC ----------------.........(((((..(((((........(--((.....))--).((((((....))))))---------...)).)))..))))).. ( -20.90, z-score = -2.24, R) >consensus ___________________UUCCGUUGUGGUG_UGGUCUUCUAAUUC_GCUAAUCGCACACGCGGCGUAUACGCAAU_____GUGCAGUACGCAUUGCCAAAUC ...........................((((........................((....)).(((....)))......................)))).... ( -7.01 = -7.09 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:10 2011