
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,709,904 – 20,710,014 |
| Length | 110 |
| Max. P | 0.730792 |

| Location | 20,709,904 – 20,710,014 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.48011 |
| G+C content | 0.39603 |
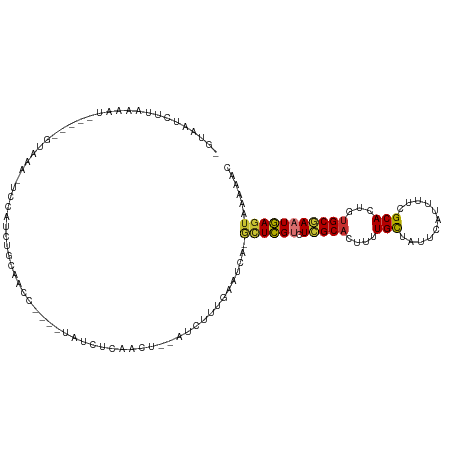
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
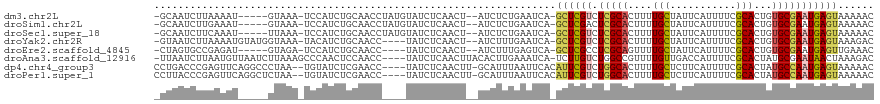
>dm3.chr2L 20709904 110 - 23011544 -GCAAUCUUAAAAU-----GUAAA-UCCAUCUGCAACCUAUGUAUCUCAACU--AUCUCUGAAUCA-GCUCGUCUCGCACUUUUGCUAUUCAUUUUCGCACUGUGCGAAUGAGUAAAAAC -............(-----(((..-......)))).................--............-((((((.((((((...(((...........)))..))))))))))))...... ( -19.00, z-score = -2.02, R) >droSim1.chr2L 20307526 110 - 22036055 -GCAAUCUUGAAAU-----GUAAA-UCCAUCUGCAACCUAUGUAUCUCAACU--AUCUCUGAAUCA-GCUCGACUCGCACUUUUGCUAUUCAUUUUCGCACUGUGCGAAUGAGUAAAAAC -.......(((..(-----(((..-......))))...........(((...--.....))).)))-(((((..((((((...(((...........)))..)))))).)))))...... ( -17.60, z-score = -0.70, R) >droSec1.super_18 629081 110 - 1342155 -GCAAUCUUCAAAU-----UUAAA-UCCAUCUGCAACCUAUGUAUCUCAACU--AUCUCUGAAUCA-GCUCGUCUCGCACUUUUGCUAUUCAUUUUCGCACUGUGCGAAUGAGUAAAAAC -.............-----.....-......((((.....))))........--............-((((((.((((((...(((...........)))..))))))))))))...... ( -18.40, z-score = -2.08, R) >droYak2.chr2R 11292877 111 + 21139217 -GUAAUCUUAAAAUGUAUGGUAAA-UACAUCUGCAACC----UAUCUCAACU--AUCUUUGAAUCA-GCUCGUCUCGCACUUUUGCUAUUCAUUUUCGCACUGUGCGAAUGAGUAAAGAC -....((((...((((((.....)-)))))........----....((((..--....))))....-((((((.((((((...(((...........)))..)))))))))))).)))). ( -23.20, z-score = -2.30, R) >droEre2.scaffold_4845 18835707 106 + 22589142 -CUAGUGCCGAGAU-----GUAGA-UCCAUCUGCAACC----UAUCUCAACU--AUCUUUGAGUCA-GCUCGCCUCGCAGUUUUGCUAUUCAUUUUCGCACUGUGCGAAUGAGUUGAAAC -.((((...(((((-----(((((-....))))))...----..)))).)))--)........(((-(((((..(((((.(..(((...........)))..)))))).))))))))... ( -27.60, z-score = -1.45, R) >droAna3.scaffold_12916 7215282 114 - 16180835 -UUAAUCUUAAUGUUAAUCUUAAAGCCCAACUCCAACC----UAUCUCAACUUACACUUGAAAUCA-UCUUGUCUGGCCGUUUUGUUGACCAUUUUCGCACUAUGCGAAUAACUAAAGAC -....((((...((((((...(..(((..((.......----....((((.......)))).....-....))..)))..)...))))))....(((((.....)))))......)))). ( -14.15, z-score = -0.93, R) >dp4.chr4_group3 4106990 113 + 11692001 CCUGACCCGAGUUCAGGCCCUAA--UGUAUCUCGAACC----UAUCUCAACUU-GCAUUUAAUUCACAUUCGUCUGGCACUUUUGCUCUUCAUUUUCGCACUAUGCCAAUGAGUAAAAAC (((((.......)))))....((--((((....((...----....))....)-)))))........((((((.(((((....(((...........)))...)))))))))))...... ( -18.60, z-score = -0.81, R) >droPer1.super_1 1248436 113 + 10282868 CCUUACCCGAGUUCAGGCUCUAA--UGUAUCUCGAACC----UAUCUCAACUU-GCAUUUAAUUCACAUUCGUCUGGCACUUUUGCUCUUCAUUUUCGCACUAUGCCAAUGAGUAAAAAC ........(((...(((.((...--........)).))----)..))).....-.............((((((.(((((....(((...........)))...)))))))))))...... ( -17.60, z-score = -0.96, R) >consensus _GUAAUCUUAAAAU_____GUAAA_UCCAUCUGCAACC____UAUCUCAACU__AUCUUUGAAUCA_GCUCGUCUCGCACUUUUGCUAUUCAUUUUCGCACUGUGCGAAUGAGUAAAAAC ...................................................................((((((.(((((....(((...........)))...)))))))))))...... (-10.80 = -10.60 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:09 2011