| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,665,068 – 20,665,168 |
| Length | 100 |
| Max. P | 0.960957 |

| Location | 20,665,068 – 20,665,168 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 61.50 |
| Shannon entropy | 0.76421 |
| G+C content | 0.35289 |
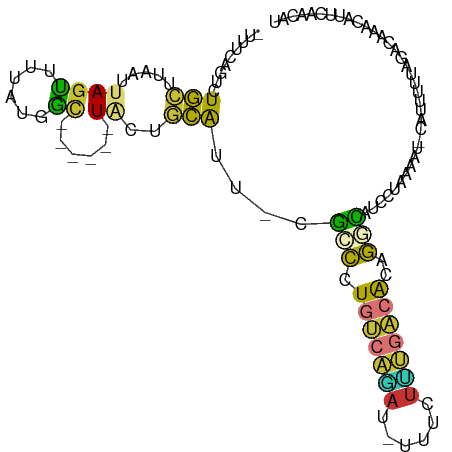
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -6.29 |
| Energy contribution | -6.03 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
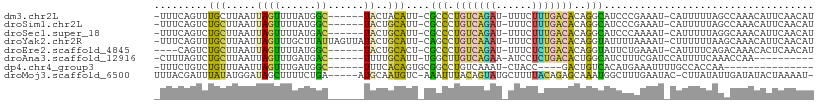
>dm3.chr2L 20665068 100 - 23011544 -UUUCAGUUUGCUUAAUUAGUUUUAUGGC------UACUACAUU-CGCCCUGUCAGAU-UUUCUUUGACACAGGCAUCCCGAAAU-CAUUUUUAGCCAAACAUUCAACAU -.....(((((((.....))).....(((------((.......-.(((.(((((((.-....)))))))..))).....(....-).....)))))))))......... ( -18.40, z-score = -2.51, R) >droSim1.chr2L 20264981 100 - 22036055 -UUUCAGUCUGCUUAAUUAGUUUUAUGGC------UACUGCAUU-CGCCCUGUCAGAU-UUUCUUUGACACAGGCAUCCCGAAAU-CAUUUUUAGCCAAACAUUCAACAU -..................(((...((((------((.......-.(((.(((((((.-....)))))))..))).....(....-).....)))))))))......... ( -18.20, z-score = -1.60, R) >droSec1.super_18 584862 100 - 1342155 -UUUCAGUCUGCUUAAUUAGUUUUAUGAC------UACUGCAUU-CGCCCUGUCAGAU-UUUCUUUGACACAGGCAUCCCAAAAU-CAUUUUUAGGCAAACAUUCAACAU -.....((.(((((((.(((((....)))------)).......-.(((.(((((((.-....)))))))..)))..........-.....))))))).))......... ( -21.90, z-score = -3.34, R) >droYak2.chr2R 11248881 106 + 21139217 -UUUCAGUUUGCUUAAUUAGUUUGCUUAUUAGUUAUACUGCAUU-CAGCCUGUCAAAU-UUUCUUUGACACAGGUAUUUUAAAAU-CUUUUUUAAGCAAACAUUCAACAU -.....((((((((((.((((..(((....)))...))))....-..((((((((((.-....))))))..))))..........-.....))))))))))......... ( -23.60, z-score = -3.87, R) >droEre2.scaffold_4845 18794206 97 + 22589142 ----CAGUCUGCUUAAUUAGUUUUAUGGC------UACUGCACU-CGCCCUGUCAGAU-UUUCUCUGACACAGGUAUUCUGAAAU-CAUUUUCAGACAAACACUCAACAU ----((((..((((((......))).)))------.))))....-.(((.(((((((.-....)))))))..)))..(((((((.-...))))))).............. ( -22.70, z-score = -2.66, R) >droAna3.scaffold_12916 7171006 91 - 16180835 -CUUUAGUCUGCUUAAUUAGUUUGAUGAC------UUUUGCAUU-UGGCUUGUCAGAA-AUCCUCUGACACUGGCAUCUUUCGAUCCAUUUUCAAACCAA---------- -........(((......((((....)))------)...))).(-(((..(((((((.-....))))))).(((.(((....))))))........))))---------- ( -17.50, z-score = -1.15, R) >dp4.chr4_group3 4047842 83 + 11692001 -UUUCUGUCUGUUUAAUUAGUUUGAUGGC------UUUCACAGUGCGGCCUGUCAAAU-CUACC----GACUGUCACAUGAAAUUUUGCCACCAA--------------- -....................(((.((((------.......(((.(((..(((....-.....----))).))).)))........)))).)))--------------- ( -14.36, z-score = 0.15, R) >droMoj3.scaffold_6500 9787849 102 + 32352404 UUUACGAUUUAUAUGGAUAGCUUUUCUGA-----AUGCAAUGUC-AAAUUUACAGUAUGCUUUUACAGAGCAAAUGGCUUUGAAUAC-CUUAUAUUGAUAUACUAAAAU- .........(((((.((((.......(((-----(.(((.(((.-......)))...))).))))((((((.....)))))).....-....)))).))))).......- ( -15.10, z-score = -0.05, R) >consensus _UUUCAGUCUGCUUAAUUAGUUUUAUGGC______UACUGCAUU_CGCCCUGUCAGAU_UUUCUUUGACACAGGCAUCCUAAAAU_CAUUUUUAGACAAACAUUCAACAU ..............................................(((.(((((((......)))))))..)))................................... ( -6.29 = -6.03 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:03 2011