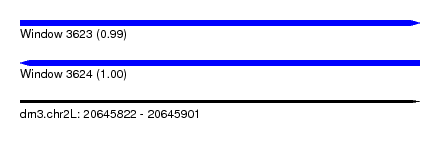
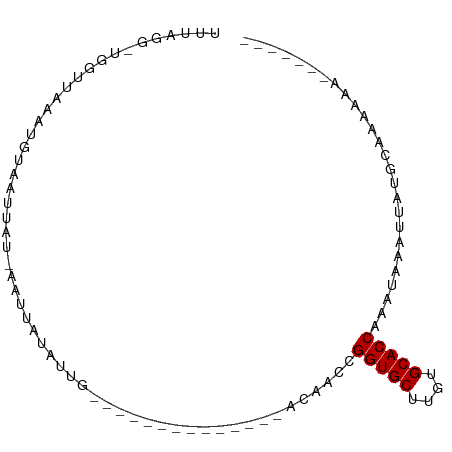
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,645,822 – 20,645,901 |
| Length | 79 |
| Max. P | 0.998384 |

| Location | 20,645,822 – 20,645,901 |
|---|---|
| Length | 79 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 68.82 |
| Shannon entropy | 0.53799 |
| G+C content | 0.29530 |
| Mean single sequence MFE | -12.64 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20645822 79 + 23011544 UUCAGUAUUUUUUGCAGAGUUUAUUUGGUGCACAAGCACCGGUAGU--------------CAAUAUAAUU-AUAAUUACAUUAAACCA-CCUAAA ....(((.....))).(.(((((..((((((....))))))(((((--------------..(((....)-)).)))))..)))))).-...... ( -13.60, z-score = -1.62, R) >droSim1.chr2L 20245340 79 + 22036055 UUCAGUAUUUUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGU--------------CAAUAUAAUU-AUAAUUACAUUAAACCA-CCUAAA (((.(((.....))).))).......(((((....)))))(((.((--------------.(((.((((.-...)))).)))..)).)-)).... ( -12.50, z-score = -1.39, R) >droSec1.super_18 565099 79 + 1342155 UUCAGUAUUUUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGU--------------CAAUAUAAUU-AUAAUUACAUUAAACCA-CCUAAA (((.(((.....))).))).......(((((....)))))(((.((--------------.(((.((((.-...)))).)))..)).)-)).... ( -12.50, z-score = -1.39, R) >droYak2.chr2R 6988645 80 + 21139217 UUCAGUAUUUUUCGCAGAAUUUAUUUGGUGCACUAGCACCGGUUGU--------------CAAUAUAAUU-AUAAUUACAUUAAACCCUCCUAAA ..........................(((((....)))))((((..--------------.(((.((((.-...)))).))).))))........ ( -11.40, z-score = -0.99, R) >droAna3.scaffold_12916 7151735 71 + 16180835 --------CAUCACCGUUCACUAUUUGGUGCACAAGCACCGGUUGC--------------CAAUAUAAUU-AUAAUUACUUUUACACA-AAUAAA --------....((((....).....(((((....))))))))...--------------..........-.................-...... ( -9.20, z-score = -0.70, R) >dp4.chr4_group3 4027796 66 - 11692001 --------------CGUAAAUUAUUUGGUGCACAAGCACCGGUUGC--------------CAAUACAAUUUAUAAAUAUUUUUACACA-AAUAAA --------------.(((((.((((((((((....)))))(((((.--------------.....)))))...)))))..)))))...-...... ( -13.70, z-score = -2.35, R) >droPer1.super_1 1172322 66 - 10282868 --------------CGUAAAUUAUUUGGUGCACAAGCACCGGUUGC--------------CAAUACAAUUUAUAAAUAUUUUUACACA-AAUAAA --------------.(((((.((((((((((....)))))(((((.--------------.....)))))...)))))..)))))...-...... ( -13.70, z-score = -2.35, R) >droGri2.scaffold_15252 234391 88 + 17193109 -------UUGAGUAUUUUGGUAUUUUGGUGCACAAGCACCGGUAAUAAUCUUUUUUUAUACAAUACACAUUAUAAUUCUAUAUAAAUAAUCCAAA -------..((((((..(((((((.((((((....))))))(((.(((.......))))))))))).))..)).))))................. ( -14.50, z-score = -1.60, R) >consensus _______UUUUUUGCAGAAUUUAUUUGGUGCACAAGCACCGGUAGU______________CAAUAUAAUU_AUAAUUACAUUAAAACA_CCUAAA ........................(((((((....)))))))..................................................... ( -9.10 = -9.10 + -0.00)

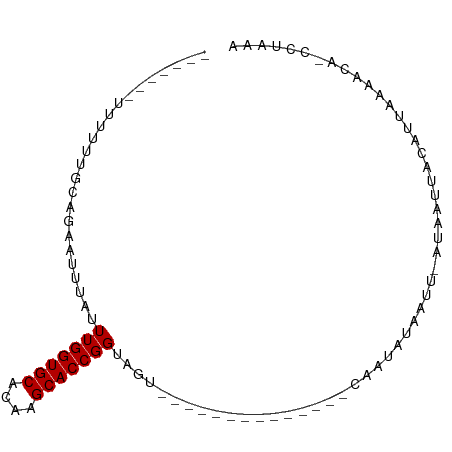
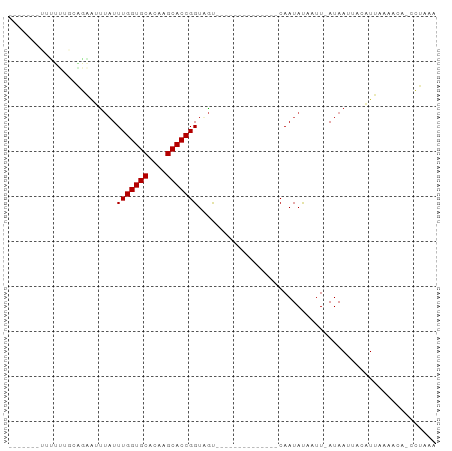
| Location | 20,645,822 – 20,645,901 |
|---|---|
| Length | 79 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 68.82 |
| Shannon entropy | 0.53799 |
| G+C content | 0.29530 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -9.40 |
| Energy contribution | -9.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
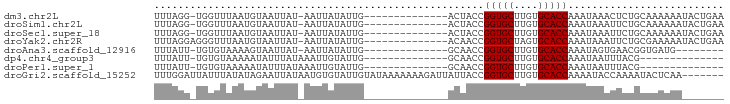
>dm3.chr2L 20645822 79 - 23011544 UUUAGG-UGGUUUAAUGUAAUUAU-AAUUAUAUUG--------------ACUACCGGUGCUUGUGCACCAAAUAAACUCUGCAAAAAAUACUGAA ....((-(((((.((((((((...-.)))))))))--------------))))))(((((....))))).......................... ( -23.00, z-score = -4.68, R) >droSim1.chr2L 20245340 79 - 22036055 UUUAGG-UGGUUUAAUGUAAUUAU-AAUUAUAUUG--------------ACUACCGGUGCUUGUGCACCAAAUAAAUUCUGCAAAAAAUACUGAA ....((-(((((.((((((((...-.)))))))))--------------))))))(((((....))))).......................... ( -23.00, z-score = -4.58, R) >droSec1.super_18 565099 79 - 1342155 UUUAGG-UGGUUUAAUGUAAUUAU-AAUUAUAUUG--------------ACUACCGGUGCUUGUGCACCAAAUAAAUUCUGCAAAAAAUACUGAA ....((-(((((.((((((((...-.)))))))))--------------))))))(((((....))))).......................... ( -23.00, z-score = -4.58, R) >droYak2.chr2R 6988645 80 - 21139217 UUUAGGAGGGUUUAAUGUAAUUAU-AAUUAUAUUG--------------ACAACCGGUGCUAGUGCACCAAAUAAAUUCUGCGAAAAAUACUGAA ..(((((.(((((((((((((...-.)))))))))--------------)..)))(((((....))))).......))))).............. ( -19.60, z-score = -2.99, R) >droAna3.scaffold_12916 7151735 71 - 16180835 UUUAUU-UGUGUAAAAGUAAUUAU-AAUUAUAUUG--------------GCAACCGGUGCUUGUGCACCAAAUAGUGAACGGUGAUG-------- ......-..(((.((.(((((...-.))))).)).--------------)))((((((((....))))).....(....))))....-------- ( -14.50, z-score = -0.76, R) >dp4.chr4_group3 4027796 66 + 11692001 UUUAUU-UGUGUAAAAAUAUUUAUAAAUUGUAUUG--------------GCAACCGGUGCUUGUGCACCAAAUAAUUUACG-------------- ..((((-(......)))))....((((((((...(--------------....).(((((....)))))..))))))))..-------------- ( -13.70, z-score = -1.51, R) >droPer1.super_1 1172322 66 + 10282868 UUUAUU-UGUGUAAAAAUAUUUAUAAAUUGUAUUG--------------GCAACCGGUGCUUGUGCACCAAAUAAUUUACG-------------- ..((((-(......)))))....((((((((...(--------------....).(((((....)))))..))))))))..-------------- ( -13.70, z-score = -1.51, R) >droGri2.scaffold_15252 234391 88 - 17193109 UUUGGAUUAUUUAUAUAGAAUUAUAAUGUGUAUUGUAUAAAAAAAGAUUAUUACCGGUGCUUGUGCACCAAAAUACCAAAAUACUCAA------- (((((....(((((((((.((........)).)))))))))..............(((((....)))))......)))))........------- ( -17.90, z-score = -2.36, R) >consensus UUUAGG_UGGUUAAAUGUAAUUAU_AAUUAUAUUG______________ACAACCGGUGCUUGUGCACCAAAUAAAUUAUGCAAAAAA_______ .......................................................(((((....))))).......................... ( -9.40 = -9.40 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:55:02 2011