
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,936,766 – 1,936,856 |
| Length | 90 |
| Max. P | 0.622159 |

| Location | 1,936,766 – 1,936,856 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 63.39 |
| Shannon entropy | 0.73327 |
| G+C content | 0.59863 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -10.06 |
| Energy contribution | -8.26 |
| Covariance contribution | -1.80 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
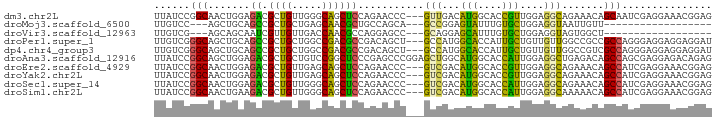
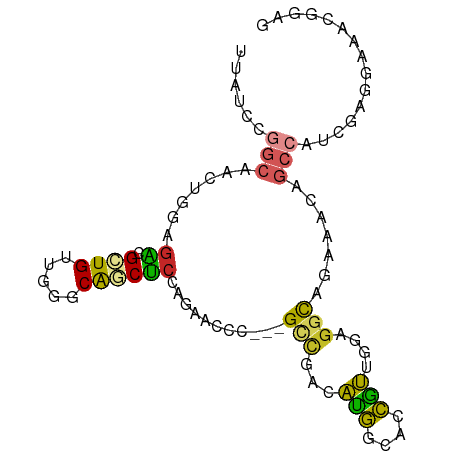
>dm3.chr2L 1936766 90 - 23011544 UUAUCCGGCAACUGGAGACGCUGUUGGGCAGCUCCAGAACCC---GUUGACAUGGCACCGUUGGAGGCAGAAACAGCAAUCGAGGAAACGGAG ...((((....((.((...((((((..((..((((((...((---((....)))).....))))))))...))))))..)).))....)))). ( -30.90, z-score = -1.80, R) >droMoj3.scaffold_6500 7383827 69 - 32352404 UUGUCC---AGCUGCAGCCGCUGCUGAGCAACGCUGCCAGCA---GCCGGAGUAUUUGUGCUGGAGGUAAUUGUU------------------ .(.(((---(((.(((((((((((((.(((....))))))))---).))).....)))))))))).)........------------------ ( -29.30, z-score = -1.78, R) >droVir3.scaffold_12963 7516643 69 + 20206255 UUGUCG---AGCAGCAAUCGUUGUUGACCAACGCCAGGAGCC---GCAGGAGCAUUUGUGCUGGAGGUAGUGGCU------------------ ..(((.---((((((....)))))))))....((((...(((---.(((..((....)).)))..)))..)))).------------------ ( -20.80, z-score = 0.14, R) >droPer1.super_1 10147695 90 - 10282868 UUGUCGGGCAGCUGCAGCCGCUGCUGGCCGACGCCGACAGCU---GCCAUGGCACCAUUGCUGUUGUUGGCCGCCGCCAGGGAGGAGGAGGAU ..(((((.((((.((....)).)))).)))))(((((((((.---((.(((....))).)).)))))))))(.((.((.....)).)).)... ( -45.20, z-score = -1.55, R) >dp4.chr4_group3 8695142 90 - 11692001 UUGUCGGGCAGCUGCAGCCGCUGCUGGCCGACGCCGACAGCU---GCCAUGGCACCAUUGCUGUUGUUGGCCGUCGCCAGGGAGGAGGAGGAU ..(((((.((((.((....)).)))).)))))(((((((((.---((.(((....))).)).)))))))))(.((.((.....))..)).).. ( -44.10, z-score = -1.50, R) >droAna3.scaffold_12916 531037 93 + 16180835 UUAUCCGGCAGCUGGAGACGCUGCUGUCCGGCUCCCGAGCCCGGAGCUGGCAUGGCACCAUUGGAGGCUGAGACAGCCAGCGAGGAGACAGAG .....(((((((.(....))))))))(((((((....)).)))))((((((.((.((((......)).))...))))))))..(....).... ( -40.70, z-score = -1.58, R) >droEre2.scaffold_4929 1982256 90 - 26641161 UUAUCCGGCAACUGGAGACGCUGUUGAGCAGCUCCAGAACCC---GUCGACAUGGCACCGUUGGAGGCAGAAACAGCCAUCGAGGAAACGGAG ...((((((..((((((..(((....)))..))))))...((---..((((........))))..))........))).....(....)))). ( -31.20, z-score = -1.91, R) >droYak2.chr2L 1917000 90 - 22324452 UUAUCCGGCAACUGGAGACGCUGUUGAGCAGCUCCAGAACCC---GUCGACAUGGCACCGUUGGAGGCAGAAACAGCCAUCGAGGAAACGGAG ...((((((..((((((..(((....)))..))))))...((---..((((........))))..))........))).....(....)))). ( -31.20, z-score = -1.91, R) >droSec1.super_14 1880021 90 - 2068291 UUAUCCGGCAACUGGAGACGCUGUUGGGCAGCUCCAGAACCC---GUCGACAUGGCACCAUUGGAGGCAGAAACAGCCAUCGAGGAAACGGAG ...((((((..((((((..(((....)))..)))))).....---(((..((((....)).))..))).......))).....(....)))). ( -31.20, z-score = -1.89, R) >droSim1.chr2L 1904207 90 - 22036055 UUAUCCGGCAACUGAAGACGCUGUUGGGCAGCUCCAGAACCC---GUCGACAUGGCACCAUUGGAGGCAAAAACAGCCAUCGAGGAAACGGAG ...((((((...((..((((...(((((....)))))....)---)))..))..)).((.(((..(((.......)))..)))))...)))). ( -26.90, z-score = -1.04, R) >consensus UUAUCCGGCAACUGGAGACGCUGUUGGGCAGCUCCAGAACCC___GCCGACAUGGCACCGUUGGAGGCAGAAACAGCCAUCGAGGAAACGGAG ......(((.......((.((((.....))))))...........(((...(((....)))....))).......)))............... (-10.06 = -8.26 + -1.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:40 2011