| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,627,745 – 20,627,846 |
| Length | 101 |
| Max. P | 0.789409 |

| Location | 20,627,745 – 20,627,846 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.78 |
| Shannon entropy | 0.52285 |
| G+C content | 0.36827 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -6.72 |
| Energy contribution | -7.47 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
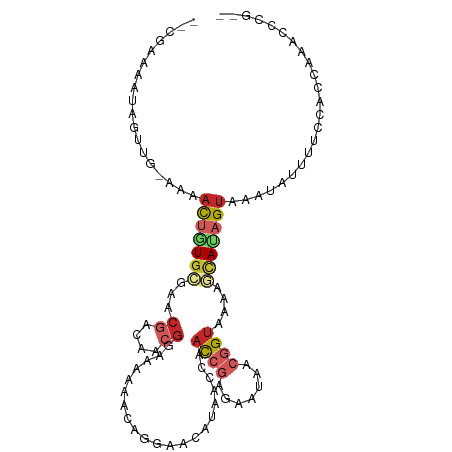
>dm3.chr2L 20627745 101 + 23011544 --CGAAAAUAGUUG-AAAACUGUGCGAACGACAACGGAAAAAACAGGAACAUAACCAACCGAGAAUACCGGUUAAAGCACAGUAAAUAUUUUCCACCAAACCCG-- --.(((((((....-...(((((((....................((.......))(((((.......)))))...)))))))...)))))))...........-- ( -20.50, z-score = -3.12, R) >droSim1.chr2L 20190860 101 + 22036055 --CGAAAAUAGUUG-AAAACUGUGCGAACGACAACGGAAAAAACAGGAACAUAACCAACCGAGAAUACCGGUUAAAGCACAGUAAAUAUUUUCCAGCAAACCCG-- --.(((((((....-...(((((((....................((.......))(((((.......)))))...)))))))...)))))))...........-- ( -20.50, z-score = -2.97, R) >droSec1.super_18 507725 101 + 1342155 --CGAAAAUAGUUG-AAAACUGUGCGAACGACAACGGAAAAAACAGGAACAUAACCAACCGAGAAUACCGGUUAAAGCACAGUAAAUAUUUUCCACCAAACCCG-- --.(((((((....-...(((((((....................((.......))(((((.......)))))...)))))))...)))))))...........-- ( -20.50, z-score = -3.12, R) >droYak2.chr2R 6975510 101 + 21139217 --CGAAAAUAGUUG-AAAACUGUGCGAGCGACAACGGAAAAAACAGGAACAUAACCAACCGAGAAUACCGGUUAAAGCAUAGUAAAUAUUUUCCACCGAGCCCG-- --.(((((((....-...(((((((....................((.......))(((((.......)))))...)))))))...)))))))...........-- ( -18.60, z-score = -1.71, R) >droEre2.scaffold_4845 18761949 101 - 22589142 --CGAAAAUAGUUG-AAAACUGUGCGAGCGACAACGGAAAAAACAGGAACAUAACCAACCGAGAAUACCGGUUAAAGCACUGUAAAUAUUUUCCGCCAAACGCG-- --((...(((((..-...))))).)).(((....(((((((.((((.....(((((.............))))).....)))).....))))))).....))).-- ( -20.62, z-score = -1.75, R) >droAna3.scaffold_12916 7129651 97 + 16180835 --CGAAAAUAGUUG-AAAACUAUGGUAGCGACAACGGAAAAAACAGGAACAUAACCAACCGAAAAUAACGGUAAAACUAUAGUAAAUAUUUUUCACCAAA------ --.(((((..(((.-...((((((((...................((.......)).((((.......))))...)))))))).)))..)))))......------ ( -15.00, z-score = -1.82, R) >droPer1.super_1 9120700 106 + 10282868 CGGAAAAAUAGUUGGAAAACUGUGCGAACGGCAACGGAAAAAACAGGAACAUAACCAACCGAGAAUAACGGUAAAACUAUAGUAAAUAUUUUUCACAACACAACAA ..((((((((((((.....((((.....((....))......)))).....))))..((((.......))))..............))))))))............ ( -20.30, z-score = -2.85, R) >dp4.chr4_group3 7674719 100 + 11692001 ----AAAAUAGUUGGAAAACUGUGCGAACGGCAACGGAAAAAACAGGAACAUAACCAACCGAAAAUAACGGUAAAACUAUAGUAAAUAUUUUUCACAACACAAC-- ----......(((((((((((((.....((....))......))))...........((((.......)))).................))))).)))).....-- ( -19.20, z-score = -3.14, R) >droWil1.scaffold_180708 10507591 95 - 12563649 ----CCUAAAUCUAAUAAAUUUUUCGU-CAAAAGCGGAAAAAACAUGCACAUUAUCUUUACAGAAUAACGGUAAUAAUAUAGUAAAUAUGCUGCUCAACU------ ----...............((((((((-.....)))))))).....((..((((((.(((.....))).)))))).....((((....))))))......------ ( -10.30, z-score = -0.37, R) >droGri2.scaffold_15126 1992882 90 - 8399593 ------------UG-AAUCUAGUGGAUAGCUGAACGGAAAAAACAAGCACAUAAGCAAUC--AAAUAACAGUAAAACUAAAGUUAAAAUGUUGUGACGACAAAUU- ------------..-......((.(...(((..............)))............--..((((((....(((....)))....))))))..).)).....- ( -8.74, z-score = 0.63, R) >consensus __CGAAAAUAGUUG_AAAACUGUGCGAACGACAACGGAAAAAACAGGAACAUAACCAACCGAGAAUAACGGUAAAAGCAUAGUAAAUAUUUUCCACCAAACCCG__ ..................(((((((...((....)).....................((((.......))))....)))))))....................... ( -6.72 = -7.47 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:58 2011