
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,618,726 – 20,618,783 |
| Length | 57 |
| Max. P | 0.666037 |

| Location | 20,618,726 – 20,618,783 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 55.28 |
| Shannon entropy | 0.77720 |
| G+C content | 0.35370 |
| Mean single sequence MFE | -7.33 |
| Consensus MFE | -4.04 |
| Energy contribution | -3.52 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.05 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
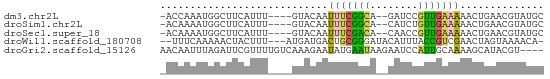
>dm3.chr2L 20618726 57 - 23011544 -ACCAAAUGGCUUCAUUU----GUACAAUUUCGGCA--GAUCCGUUGAAAAACUGAACGUAUGC -..((((((....)))))----).....(((((((.--.....))))))).............. ( -8.90, z-score = 0.04, R) >droSim1.chr2L 20181991 57 - 22036055 -ACAAAAUGGCUUCAUUU----GUACAAUUUCGGCA--CAUCUGUUGAAAAACUGAACGUAUGC -............(((.(----((.((.((((((((--....))))))))...)).))).))). ( -7.80, z-score = 0.12, R) >droSec1.super_18 498908 57 - 1342155 -ACAAAAUGGCUUCAUUU----GUACAAUUUCGACA--CAACCGUUGAAAAACUGAACGUAUGC -............(((.(----((.((.(((((((.--.....)))))))...)).))).))). ( -7.70, z-score = -0.07, R) >droWil1.scaffold_180708 2031873 58 - 12563649 --UUUCAAAAACUACUUU---AUGAUGACUGCGGGAUACAUUUACCGUCGAACUAGUAAAACA- --.............(((---((.......((((..........)))).......)))))...- ( -5.84, z-score = -0.03, R) >droGri2.scaffold_15126 6392081 60 - 8399593 AACAAUUUAGAUUCGUUUUGUCAAAGAAUAUGAAUAAGAAUCCAUUGCAAAAGCAUACGU---- .........(((((......(((.......)))....)))))...(((....))).....---- ( -6.40, z-score = -0.31, R) >consensus _ACAAAAUGGCUUCAUUU____GUACAAUUUCGGCA__CAUCCGUUGAAAAACUGAACGUAUGC ............................(((((((........))))))).............. ( -4.04 = -3.52 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:55 2011