
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,618,539 – 20,618,636 |
| Length | 97 |
| Max. P | 0.655283 |

| Location | 20,618,539 – 20,618,636 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 67.55 |
| Shannon entropy | 0.66399 |
| G+C content | 0.48707 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -10.78 |
| Energy contribution | -9.67 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
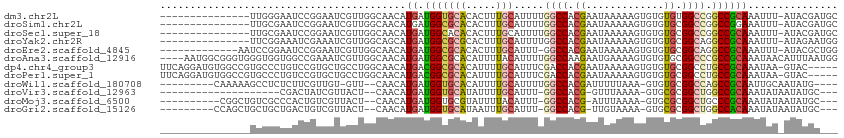
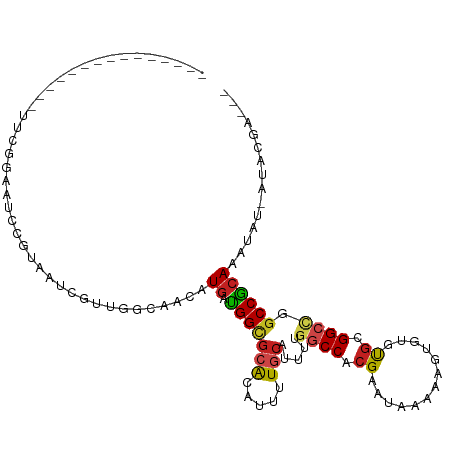
>dm3.chr2L 20618539 97 + 23011544 ---------------UUGGGAAUCCGGAAUCGUUGGCAACAUGAUGGUGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGUGGCCGGCCGCAAAUUU-AUACGAUGC ---------------(((((...((((.(((((.(....)))))).(..((((((((....((((....))))...))))))))..)..)))))).)))....-......... ( -27.70, z-score = -0.29, R) >droSim1.chr2L 20181809 97 + 22036055 ---------------UUGCGAAUCCGGAAUCGUUGGCAACAUGAUGGCGCACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGGAAAUUU-AUACGAUGC ---------------.......(((((.(((((.(....)))))).(((((((((((....((((....))))...)))))))))))......))))).....-......... ( -32.70, z-score = -1.51, R) >droSec1.super_18 498725 97 + 1342155 ---------------UUGCGAAUCCGGAAUCGUUGGCAACAUGAUGGCACACACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGCAAAUUU-AUACGAUGC ---------------(((((...((((.(((((.(....)))))).(((((((((((....((((....))))...)))))))))))..)))).)))))....-......... ( -36.50, z-score = -3.00, R) >droYak2.chr2R 6966668 97 + 21139217 ---------------UUCGGAAAUCGAAAUCGUUGGCAGCAUGAUGGCGCGCACUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCAGGCCGCAAAUUU-AUAGAAUGG ---------------((((.....)))).......((.((.((.(.(((((((((((....((((....))))...))))))))))).))).)).))......-......... ( -27.90, z-score = -0.34, R) >droEre2.scaffold_4845 18754126 98 - 22589142 -------------AAUCCGGAAUCCGGAAUCGUUGGCAACAUGAUGGCGCACACUUUGCAUUU-GGCCACGAAUAAAAAGUGUGUGCGGCAGGCCGCAAAUUU-AUACGCUGG -------------..(((((...)))))(((((.(....))))))((((((((((((..((((-(....)))))..))))))))(((((....))))).....-...)))).. ( -36.90, z-score = -2.80, R) >droAna3.scaffold_12916 7118928 109 + 16180835 ----AAUGGCGGGUGGGUGGUGGCCGAAAUCGUUGGCAACAUGAUGGCGCACAUUUUACAUUUUGGCCAAGAAUGAAAAGUGUGUGCGGCCCGCCGCAAAUAACAUUUAAUGG ----...(((((((..(((...(((((.....)))))..)))....(((((((((((.((((((.....)))))).))))))))))).))))))).................. ( -43.90, z-score = -3.09, R) >dp4.chr4_group3 3995475 107 - 11692001 UUCAGGAUGUGGCCGUGCCCUGUCCGUGCUGCCUGGCAACAUGACGGCGCACAUUUUGCAUUUCGACCACGAAUAAAAAGUGUGUGCGGCCUGCCGCAAAUAA-GUAC----- .......((((((.(.(((.((((..((((....))))....))))(((((((((((....((((....))))...))))))))))))))).)))))).....-....----- ( -40.20, z-score = -2.06, R) >droPer1.super_1 1140593 107 - 10282868 UUCAGGAUGUGGCCGUGCCCUGUCCGUGCUGCCUGGCAACAUGACGGCGCACAUUUUGCAUUUCGACCACGAAUAAAAAGUGUGUGCGGCCUGCCGCAAAUAA-GUAC----- .......((((((.(.(((.((((..((((....))))....))))(((((((((((....((((....))))...))))))))))))))).)))))).....-....----- ( -40.20, z-score = -2.06, R) >droWil1.scaffold_180708 2031684 96 + 12563649 ---------CAAAAAGCCUCUCUUCGUUGU-GUU--CAACAUGAUGGUGCACAUUUUGCAUUUUGGCCACGAUUUUUAAA-GUGUGCGGCCAGCCGCAAUUGCAAUAUG---- ---------......(((.....((((..(-(..--...))..)))).((((((((((.(..(((....)))..).))))-))))))))).....((....))......---- ( -25.20, z-score = -0.60, R) >droVir3.scaffold_12963 6733462 85 - 20206255 --------------------CGACUAUCGUUACU--CAACAUGAUGGUGCAUAUUUUGCAUUU-GGCCACG-GUUUAAAA-GUGCGCGGCUGGCCGCAAAUAUAAUAUGC--- --------------------..((((((((....--....))))))))(((((((..((((((-.((....-))....))-))))((((....))))......)))))))--- ( -26.00, z-score = -1.66, R) >droMoj3.scaffold_6500 24709983 95 + 32352404 ----------CGGCUGUCGCCCACUGUCGUUACU--CAACAUGAUGGUGCGUAUUUUACAUUU-GGCCACG-AUUUAAAA-GUGCGCGGCUGCCCGCAAAUAUAAUAUGC--- ----------.(((.(((((.(((((((((....--....))))))))).(((((((....((-(....))-)....)))-))))))))).)))................--- ( -27.40, z-score = -0.72, R) >droGri2.scaffold_15126 6391856 96 + 8399593 ---------CCAGCUGCUGCUGACUGUCGUUACU--CAACAUGAUGGUGCAUAAUUUGCAUUU-GGCCACG-UUGUAAAA-GUGCGCGGCUGGCCACAAAUAUAAUAUGC--- ---------.((((....))))((((((((....--....))))))))((((((((((....(-(((((.(-((((....-....))))))))))))))))....)))))--- ( -29.40, z-score = -0.86, R) >consensus _______________UUCGGAAUCCGUAAUCGUUGGCAACAUGAUGGCGCACAUUUUGCAUUUUGGCCACGAAUAAAAAGUGUGUGCGGCCGGCCGCAAAUAU_AUACGA___ ..............................................((((((.((((...................)))).))))))((....)).................. (-10.78 = -9.67 + -1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:54 2011