| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,606,241 – 20,606,332 |
| Length | 91 |
| Max. P | 0.988510 |

| Location | 20,606,241 – 20,606,331 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 54.88 |
| Shannon entropy | 0.78253 |
| G+C content | 0.36377 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -14.08 |
| Energy contribution | -12.92 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
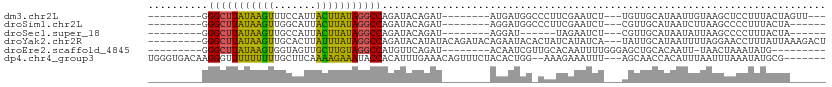
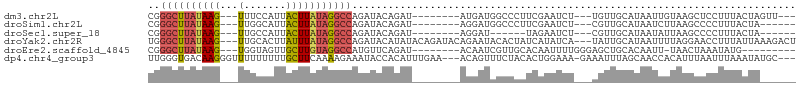
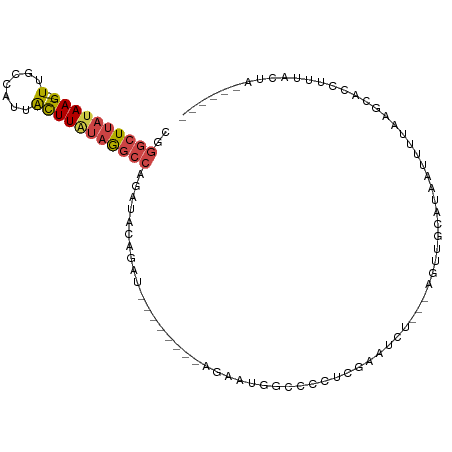
>dm3.chr2L 20606241 90 - 23011544 ---------GGGCUUAUAAGUUUCCAUUACUUAUAGGCCAGAUACAGAU--------AUGAUGGCCCUUCGAAUCU---UGUUGCAUAAUUGUAAGCUCCUUUACUAGUU--- ---------.(((((((((((.......)))))))))))......((((--------.(((.......))).))))---..(((((....)))))...............--- ( -20.20, z-score = -0.90, R) >droSim1.chr2L 20169640 87 - 22036055 ---------GGGCUUAUAAGUUGGCAUUACUUAUAGGCCAGAUACAGAU--------AGGAUGGCCCUUCGAAUCU---CGUUGCAUAAUCUUAAGCCCCUUUACUA------ ---------(((((((...(((((((.........(((((....(....--------.)..)))))...((.....---)).))).))))..)))))))........------ ( -22.00, z-score = -0.76, R) >droSec1.super_18 486776 81 - 1342155 ---------GGGCUUAUAAGUUGCCAUUACUUAUAGGCCAGAUACAGAU--------AGGAU------UAGAAUCU---CGUUGCAUAAUAUUAAGCCCCUUUACUA------ ---------.(((((((((((.......))))))))))).......((.--------.((((------....))))---..))........................------ ( -17.10, z-score = -0.65, R) >droYak2.chr2R 6954364 101 - 21139217 ---------GGGCUUAUAAGUUGCACUUAUUUAUAGGCCAGAUACAUAUACAGAUACAGAAUACACUAUCAUAUCA---UAUUGCAUAAUUUUAGGAACCUUUAUUAAAGACU ---------.(((((((((((.......))))))))))).............((((..((........)).)))).---.......((((...((....))..))))...... ( -14.50, z-score = -0.97, R) >droEre2.scaffold_4845 18741323 86 + 22589142 ---------GGGCUUAUAAGUGGUAGUUGCUUGUAGGCCAUGUUCAGAU--------ACAAUCGUUGCACAAUUUUGGGAGCUGCACAAUU-UAACUAAAUAUG--------- ---------.((((((((((..(...)..)))))))))).(((.(((..--------.(((..(((....))).)))....))).)))...-............--------- ( -20.70, z-score = -0.68, R) >dp4.chr4_group3 3978254 101 + 11692001 UGGGUGACAAGGGUUUUUUUUUGCUUCAAAAGAAAUACCACAUUUGAAACAGUUUCUACACUGG--AAAGAAAUUU---AGCAACCACAUUUAAUUUAAAUAUGCG------- ..(....)...(((.(((((((......))))))).)))..(((((((.((((......)))).--(((....)))---...............))))))).....------- ( -14.10, z-score = 0.36, R) >consensus _________GGGCUUAUAAGUUGCCAUUACUUAUAGGCCAGAUACAGAU________AGAAUGG_CCAUCAAAUCU___AGUUGCAUAAUUUUAAGCACCUUUACUA______ ..........(((((((((((.......))))))))))).......................................................................... (-14.08 = -12.92 + -1.16)
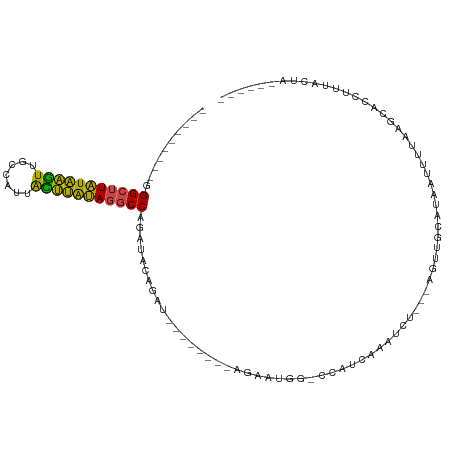
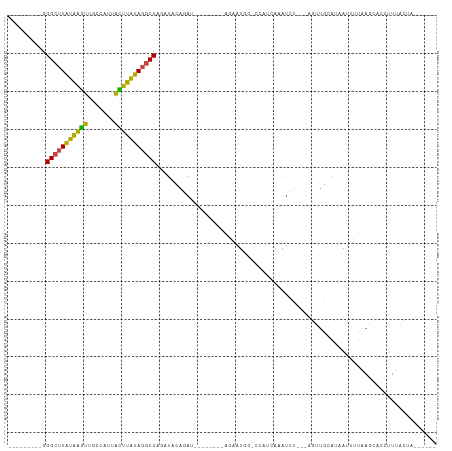
| Location | 20,606,241 – 20,606,332 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 55.75 |
| Shannon entropy | 0.81138 |
| G+C content | 0.36630 |
| Mean single sequence MFE | -18.17 |
| Consensus MFE | -7.92 |
| Energy contribution | -8.53 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20606241 91 - 23011544 CGGGCUUAUAAG---UUUCCAUUACUUAUAGGCCAGAUACAGAU--------AUGAUGGCCCUUCGAAUCU---UGUUGCAUAAUUGUAAGCUCCUUUACUAGUU--- ..((((((((((---(.......)))))))))))......((((--------.(((.......))).))))---..(((((....)))))...............--- ( -20.20, z-score = -0.84, R) >droSim1.chr2L 20169640 88 - 22036055 CGGGCUUAUAAG---UUGGCAUUACUUAUAGGCCAGAUACAGAU--------AGGAUGGCCCUUCGAAUCU---CGUUGCAUAAUCUUAAGCCCCUUUACUA------ .(((((((...(---((((((.........(((((....(....--------.)..)))))...((.....---)).))).))))..)))))))........------ ( -22.30, z-score = -0.77, R) >droSec1.super_18 486776 82 - 1342155 CGGGCUUAUAAG---UUGCCAUUACUUAUAGGCCAGAUACAGAU--------AGGAU------UAGAAUCU---CGUUGCAUAAUAUUAAGCCCCUUUACUA------ ..((((((((((---(.......))))))))))).......((.--------.((((------....))))---..))........................------ ( -17.10, z-score = -0.60, R) >droYak2.chr2R 6954364 102 - 21139217 UGGGCUUAUAAG---UUGCACUUAUUUAUAGGCCAGAUACAUAUACAGAUACAGAAUACACUAUCAUAUCA---UAUUGCAUAAUUUUAGGAACCUUUAUUAAAGACU ..((((((((((---(.......))))))))))).............((((..((........)).)))).---.......((((...((....))..))))...... ( -14.50, z-score = -0.60, R) >droEre2.scaffold_4845 18741323 87 + 22589142 CGGGCUUAUAAG---UGGUAGUUGCUUGUAGGCCAUGUUCAGAU--------ACAAUCGUUGCACAAUUUUGGGAGCUGCACAAUU-UAACUAAAUAUG--------- ..((((((((((---..(...)..)))))))))).(((.(((..--------.(((..(((....))).)))....))).)))...-............--------- ( -20.70, z-score = -0.66, R) >dp4.chr4_group3 3978255 101 + 11692001 UUGGGUGACAAGGGUUUUUUUUUGCUUCAAAAGAAAUACCACAUUUGAA---ACAGUUUCUACACUGGAAA-GAAAUUUAGCAACCACAUUUAAUUUAAAUAUGC--- .(((((..(...(((.(((((((......))))))).)))...(((...---.((((......))))....-))).....)..))).))................--- ( -14.20, z-score = 0.44, R) >consensus CGGGCUUAUAAG___UUGCCAUUACUUAUAGGCCAGAUACAGAU________AGAAUGGCCCCUCGAAUCU___AGUUGCAUAAUUUUAAGCACCUUUACUA______ ..((((((((((............)))))))))).......................................................................... ( -7.92 = -8.53 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:51 2011