| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,593,916 – 20,594,006 |
| Length | 90 |
| Max. P | 0.715661 |

| Location | 20,593,916 – 20,594,006 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 67.82 |
| Shannon entropy | 0.55011 |
| G+C content | 0.61957 |
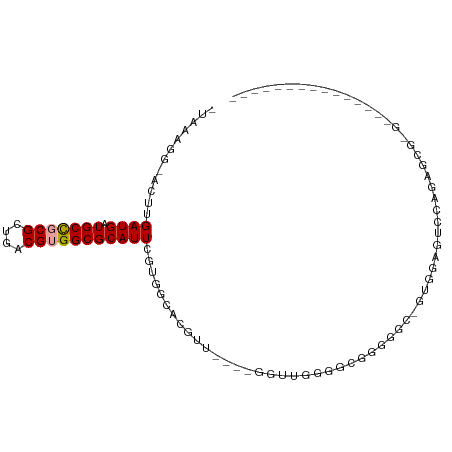
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -10.63 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.08 |
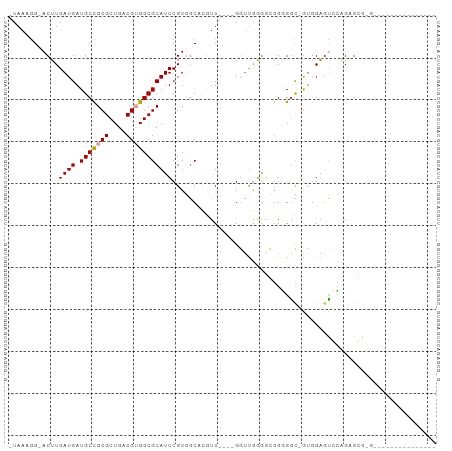
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
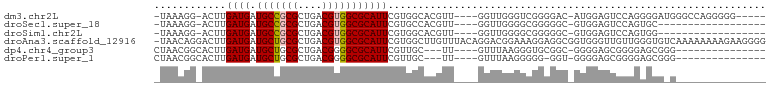
>dm3.chr2L 20593916 90 - 23011544 -UAAAGG-ACUUGAUGAUGCCGCGCUGACGUGGCGCAUUCGUGGCACGUU----GGUUGGGUCGGGGAC-AUGGAGUCCAGGGGAUGGGCCAGGGGG----- -....((-((((.(((.(.((((((..(((((.(((....))).))))).----.))...).))).).)-)).))))))..((......))......----- ( -36.10, z-score = -2.34, R) >droSec1.super_18 469630 77 - 1342155 -UAAAGG-ACUUGAUGAUGCCGCGCUGACGUGGCGCAUUCGUGCCACGUU----GGUUGGGGCGGGGGC-GUGGAGUCCAGUGC------------------ -....((-((((.(((.((((.(((..(((((((((....))))))))).----.).)).))))....)-)).)))))).....------------------ ( -43.40, z-score = -5.41, R) >droSim1.chr2L 20157439 77 - 22036055 -UAAAGG-ACUUGAUGAUGCCGCGCUGACGUGGCGCAUUCGUGGCACGUU----GGUUGGGGCGGGGGC-GUGGAGUCCAGUGG------------------ -....((-((((.(((.((((.(((..(((((.(((....))).))))).----.).)).))))....)-)).)))))).....------------------ ( -38.80, z-score = -4.61, R) >droAna3.scaffold_12916 7089512 101 - 16180835 -UAACAGGACUUGAUGAUGCUGCGCUGACGUGGCGCAUUCGUGGCUUGUUUACAGGACGGAAAGGAGGCGGUGGGUUGUUGGGUGUCAAAAAAAAGAAGGGG -.....(((((..(..((.(((((((.....))))))(((((..((((....)))))))))...........).))..)..))).))............... ( -24.10, z-score = 0.08, R) >dp4.chr4_group3 3962577 79 + 11692001 CUAACGGCACUUGAUGAUGCUGCGCUGACGGGGCGCAUUCGUUGC---UU----GUUUAAGGGUGCGGC-GGGGAGCGGGGAGCGGG--------------- ....(.((.((((....((((((((((((((((((....)))..)---))----)))....))))))))-).....))))..)).).--------------- ( -25.50, z-score = -0.09, R) >droPer1.super_1 1108045 78 + 10282868 CUAACGGCACUUGAUGAUGCUGCGCUGACGGGGCGCAUUCGUUGC---UU----GUUUAAGGGGG-GGU-GGGGAGCGGGGAGCGGG--------------- ....(.((.(((((((.((((.((....)).))))))))((((.(---(.----.(((......)-)).-.)).))))))).)).).--------------- ( -22.30, z-score = -0.07, R) >consensus _UAAAGG_ACUUGAUGAUGCCGCGCUGACGUGGCGCAUUCGUGGCACGUU____GGUUGGGGCGGGGGC_GUGGAGUCCAGAGCG_G_______________ ............((((.(((((((....)))))))))))............................................................... (-10.63 = -10.72 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:49 2011