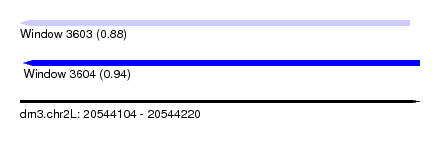
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,544,104 – 20,544,220 |
| Length | 116 |
| Max. P | 0.936695 |

| Location | 20,544,104 – 20,544,217 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Shannon entropy | 0.33834 |
| G+C content | 0.42080 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20544104 113 - 23011544 --CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCC-GCCGCC-- --....(((..((((.(((.....)))(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))))..)))...(((.-...)))-- ( -29.80, z-score = -1.12, R) >droSim1.chr2L 20114801 113 - 22036055 --CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCC-GCCGCC-- --....(((..((((.(((.....)))(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))))..)))...(((.-...)))-- ( -29.80, z-score = -1.12, R) >droSec1.super_18 426380 113 - 1342155 --CCGCGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCC-GCCGCC-- --..((((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...))))..........(((.-...)))-- ( -33.10, z-score = -1.79, R) >droYak2.chr2R 6900000 113 - 21139217 --UCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGUCC-GCCGCC-- --....((.....))............(((((....(((((((((((..(((....((.......))....)))..)))))).)))))(..((.......))..)....-))))).-- ( -28.80, z-score = -1.43, R) >droEre2.scaffold_4845 18688739 113 + 22589142 --CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUCUCGGCC-GCCGCC-- --....((.....))............(((((..(((((((((((((..(((....((.......))....)))..))))))......(.....)......))))))).-))))).-- ( -30.70, z-score = -1.28, R) >droAna3.scaffold_12916 7046213 115 - 16180835 --CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAACUCGGUCU-CCUCCCGA --...(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))........((((...-....)))) ( -30.00, z-score = -1.66, R) >droPer1.super_1 1055003 114 + 10282868 --CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAGUUCGGCCUAGGCGCC-- --...((((((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...((.((......)).))...))))..-- ( -31.00, z-score = -1.14, R) >dp4.chr4_group3 3910823 114 + 11692001 --CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAGUUCGGCCUAGGCGCC-- --...((((((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...((.((......)).))...))))..-- ( -31.00, z-score = -1.14, R) >droWil1.scaffold_180708 10696014 105 + 12563649 --------UAUAUGCAUUAUAAUUUGAGUGGCACGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGU--CCGGUCCAGUCGCCACCGCCCUC--- --------.................(((.(((...((((((((((((..(((....((.......))....)))..)))))).)))))).--..(((......)))...))))))--- ( -28.60, z-score = -2.44, R) >droVir3.scaffold_12963 6651909 101 + 20206255 ----------UAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGUGGCAGUUACCACGACUCU------- ----------(((((.(((.....)))...)))))..((((((((((..(((....((.......))....)))..)))))))))).((((((((......))))))))..------- ( -32.00, z-score = -3.78, R) >droMoj3.scaffold_6500 24622958 114 - 32352404 UCGGCCUGUGUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGCAGUUGCCACGACUCUCGG---- ..((((.((((((((.(((.....)))...))).((((((((..(........)..))))))))......))))).)))).......(((((.(((....))).))))).....---- ( -33.20, z-score = -1.58, R) >droGri2.scaffold_15126 6310325 108 - 8399593 UCGUAUUGUGUAUGCAUUAUAAUUUGAGUGGCACGCUGAGAGUAGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUCAGGC---UGCCACAUUUCC------- (((.((((((.......)))))).)))((((((((((((((((.(((..(((....((.......))....)))..))).)).)))))))....).---))))))......------- ( -27.50, z-score = -1.77, R) >consensus __CCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAGUUUCGGCC_GCCGCC__ .....(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))........................ (-22.70 = -23.16 + 0.46)

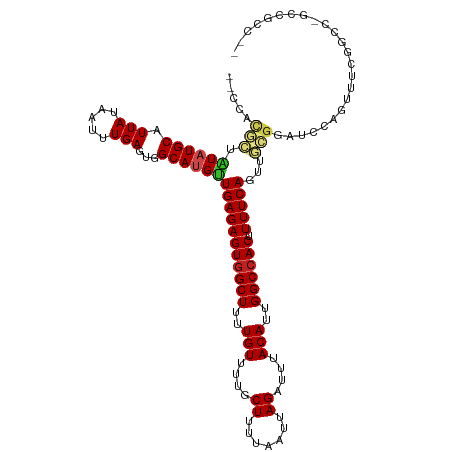
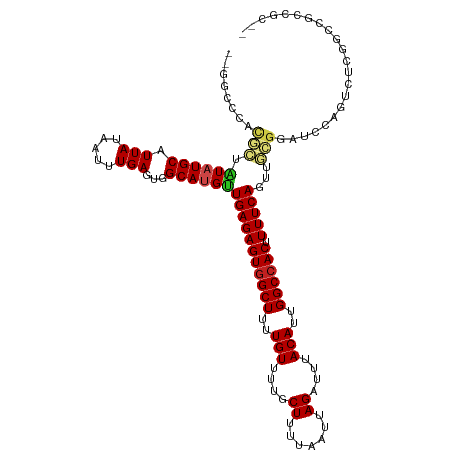
| Location | 20,544,105 – 20,544,220 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.54 |
| Shannon entropy | 0.34407 |
| G+C content | 0.42644 |
| Mean single sequence MFE | -33.19 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20544105 115 - 23011544 --GGCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGC-- --((((...(((..((((.(((.....)))(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))))..)))...)))).....-- ( -36.60, z-score = -2.58, R) >droSim1.chr2L 20114802 115 - 22036055 --GGCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGC-- --((((...(((..((((.(((.....)))(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))))..)))...)))).....-- ( -36.60, z-score = -2.58, R) >droSec1.super_18 426381 116 - 1342155 -GGGCCCGCGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGC-- -.((((...(((..((((.(((.....)))(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))))..)))...)))).....-- ( -36.60, z-score = -1.80, R) >droYak2.chr2R 6900001 115 - 21139217 --GGCUCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGUCCGCCGC-- --(((..((((.....))........(((..(((((....((((((((((..(((....((.......))....)))..))))))))))....))).))..)))....))..)))..-- ( -33.10, z-score = -2.08, R) >droEre2.scaffold_4845 18688740 115 + 22589142 --GGCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUCUCGGCCGCCGC-- --((((...(((..((((.(((.....)))(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))))..)))...)))).....-- ( -36.60, z-score = -2.40, R) >droAna3.scaffold_12916 7046217 114 - 16180835 --GGCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAA-CUCGGUCUCCUC-- --((((...((.....))..........((((.(((....((((((((((..(((....((.......))....)))..))))))))))....)))(....).)-))))))).....-- ( -32.30, z-score = -2.16, R) >droPer1.super_1 1055004 115 + 10282868 ---GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAG-UUCGGCCUAGGCGC ---(((...(((((((((.(((.....)))...)))))).((((((((((..(((....((.......))....)))..)))))))))).))).((.((.....-.)).))...))).. ( -31.80, z-score = -0.99, R) >dp4.chr4_group3 3910824 116 + 11692001 --GGCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAG-UUCGGCCUAGGCGC --((((..(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))((.....-.))))))....... ( -34.70, z-score = -1.60, R) >droWil1.scaffold_180708 10696015 104 + 12563649 -----------UAUAUGCAUUAUAAUUUGAGUGGCACGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGU--CCGGUCCAGUCGCCACCGCCCU-- -----------.................((.(((..((((((((((((((..(((....((.......))....)))..)))))).)))))).--.))..))).))...........-- ( -26.40, z-score = -1.62, R) >droVir3.scaffold_12963 6651910 100 + 20206255 -------------UAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGUGGCAGUUACCACGACUC------ -------------(((((.(((.....)))...)))))..((((((((((..(((....((.......))....)))..)))))))))).((((((((......)))))))).------ ( -32.00, z-score = -3.97, R) >droMoj3.scaffold_6500 24622962 113 - 32352404 UGCUCGGCCUGUGUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGCAGUUGCCACGACUC------ ..((((((.(((.(((.((........)).)))))).))))))(((((((..(((....((.......))....)))..)))))))....(((((.(((....))).))))).------ ( -34.00, z-score = -1.83, R) >droGri2.scaffold_15126 6310326 110 - 8399593 CACUCGUAUUGUGUAUGCAUUAUAAUUUGAGUGGCACGCUGAGAGUAGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUCAGGC---UGCCACAUUUC------ ((((((.((((((.......)))))).))))))((..(((((((((.(((..(((....((.......))....)))..))).)).)))))))....))---...........------ ( -27.60, z-score = -1.59, R) >consensus __GGCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAGUCUCGGCCGCCGC__ ........(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))...................... (-22.70 = -23.16 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:46 2011