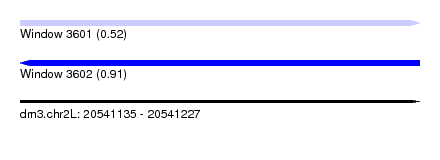
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,541,135 – 20,541,227 |
| Length | 92 |
| Max. P | 0.912634 |

| Location | 20,541,135 – 20,541,227 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.01 |
| Shannon entropy | 0.32596 |
| G+C content | 0.34826 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -12.33 |
| Energy contribution | -12.51 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
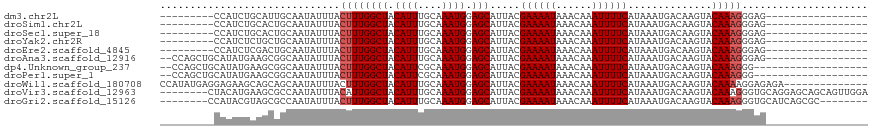
>dm3.chr2L 20541135 92 + 23011544 ---------CCAUCUGCAUUGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG----------------- ---------((...(((...))).......((((((((.((((....)))).))).....((((((......))))))..............)))))))..----------------- ( -14.90, z-score = -0.99, R) >droSim1.chr2L 20111854 92 + 22036055 ---------CCAUCUGCACUGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG----------------- ---------((...(((...))).......((((((((.((((....)))).))).....((((((......))))))..............)))))))..----------------- ( -14.90, z-score = -1.09, R) >droSec1.super_18 423451 92 + 1342155 ---------CCAUCUGCACUGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG----------------- ---------((...(((...))).......((((((((.((((....)))).))).....((((((......))))))..............)))))))..----------------- ( -14.90, z-score = -1.09, R) >droYak2.chr2R 6897123 92 + 21139217 ---------CCAUCUCUGCUGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG----------------- ---------...(((((..(((.............(((.((((....)))).))).....((((((......))))))...........)))...))))).----------------- ( -15.10, z-score = -0.99, R) >droEre2.scaffold_4845 18685803 92 - 22589142 ---------CCAUCUCGACUGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG----------------- ---------...((((.(((....((((((.....(((.((((....)))).))).....((((((......)))))).))))))...))).....)))).----------------- ( -14.50, z-score = -1.34, R) >droAna3.scaffold_12916 7043616 99 + 16180835 --CCAGCUGCAUAUGAAGCGGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG----------------- --((.(((((.......)))))........((((((((.((((....)))).))).....((((((......))))))..............)))))))..----------------- ( -22.50, z-score = -2.85, R) >dp4.Unknown_group_237 11005 97 - 11213 --CCAGCUGCAUAUGAAGCGGCAAUAUUUACUUUGGCUACAUUCGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGG------------------- --((.(((((.......)))))........((((((((.((((....)))).))).....((((((......))))))..............)))))))------------------- ( -21.30, z-score = -2.71, R) >droPer1.super_1 1050976 97 - 10282868 --CCAGCUGCAUAUGAAGCGGCAAUAUUUACUUUGGCUACAUUCGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGG------------------- --((.(((((.......)))))........((((((((.((((....)))).))).....((((((......))))))..............)))))))------------------- ( -21.30, z-score = -2.71, R) >droWil1.scaffold_180708 10692345 104 - 12563649 CCAUAUGAGGAGAAGCAGCAGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAAGGAGAGA-------------- ((............((....))......(((((.((((.((((....)))).))).....((((((......))))))........)))))).....)).....-------------- ( -13.30, z-score = 0.10, R) >droVir3.scaffold_12963 6648869 110 - 20206255 --------CUACAUGAAGCGCCAAUAUUUACAUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGUGCAGGAGCAGCAGUUGGA --------.(((.((..((((((((......))))))..((((....))))..)).....((((((......))))))........)).)))(((..(.(((....))).)..))).. ( -22.70, z-score = -1.23, R) >droGri2.scaffold_15126 6307302 102 + 8399593 --------CCAUACGUAGCGCCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGUGCAUCAGCGC-------- --------.........(((((......(((((.((((.((((....)))).))).....((((((......))))))........)))))).....)))))........-------- ( -20.00, z-score = -0.95, R) >consensus _________CAAACGAAGCGGCAAUAUUUACUUUGGCUACAUUUGCAAAUGGAGCAUUACGAAAAUAAACAAAUUUUCAUAAAUGACAAGUACAAAGGGAG_________________ ..............................((((((((.((((....)))).))).....((((((......))))))..............)))))..................... (-12.33 = -12.51 + 0.18)
| Location | 20,541,135 – 20,541,227 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.01 |
| Shannon entropy | 0.32596 |
| G+C content | 0.34826 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.66 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20541135 92 - 23011544 -----------------CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCAAUGCAGAUGG--------- -----------------...................((((((((((......))))))))))....(((((((((..(((((............))))).)))))))))--------- ( -18.70, z-score = -1.95, R) >droSim1.chr2L 20111854 92 - 22036055 -----------------CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCAGUGCAGAUGG--------- -----------------..((.((((((((.((...((((((((((......)))))))))).(((.((((....)))).))).............)))))))))).))--------- ( -21.00, z-score = -2.63, R) >droSec1.super_18 423451 92 - 1342155 -----------------CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCAGUGCAGAUGG--------- -----------------..((.((((((((.((...((((((((((......)))))))))).(((.((((....)))).))).............)))))))))).))--------- ( -21.00, z-score = -2.63, R) >droYak2.chr2R 6897123 92 - 21139217 -----------------CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCAGCAGAGAUGG--------- -----------------....((((((.........((((((((((......))))))))))(((...((((((.............))))))...)))))))))....--------- ( -18.02, z-score = -1.41, R) >droEre2.scaffold_4845 18685803 92 + 22589142 -----------------CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCAGUCGAGAUGG--------- -----------------...........((((.(..((((((((((......))))))))))(((...((((((.............))))))...)))).))))....--------- ( -16.52, z-score = -1.35, R) >droAna3.scaffold_12916 7043616 99 - 16180835 -----------------CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCCGCUUCAUAUGCAGCUGG-- -----------------...................((((((((((......))))))))))....(((.(((((.(((.(((...(((....)))...))).))).))))).)))-- ( -20.10, z-score = -2.03, R) >dp4.Unknown_group_237 11005 97 + 11213 -------------------CCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCGAAUGUAGCCAAAGUAAAUAUUGCCGCUUCAUAUGCAGCUGG-- -------------------.................((((((((((......))))))))))....(((.(((((.(((.(((...(((....)))...))).))).))))).)))-- ( -19.40, z-score = -1.52, R) >droPer1.super_1 1050976 97 + 10282868 -------------------CCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCGAAUGUAGCCAAAGUAAAUAUUGCCGCUUCAUAUGCAGCUGG-- -------------------.................((((((((((......))))))))))....(((.(((((.(((.(((...(((....)))...))).))).))))).)))-- ( -19.40, z-score = -1.52, R) >droWil1.scaffold_180708 10692345 104 + 12563649 --------------UCUCUCCUUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCUGCUGCUUCUCCUCAUAUGG --------------........((((((..((....((((((((((......)))))))))).....))..)))))).(((((...((((.....))))))))).............. ( -18.90, z-score = -2.73, R) >droVir3.scaffold_12963 6648869 110 + 20206255 UCCAACUGCUGCUCCUGCACCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAUGUAAAUAUUGGCGCUUCAUGUAG-------- .....((((.((...((((...((((((..((....((((((((((......)))))))))).....))..))))))))))(((((((....))))))))).....))))-------- ( -25.80, z-score = -3.25, R) >droGri2.scaffold_15126 6307302 102 - 8399593 --------GCGCUGAUGCACCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGGCGCUACGUAUGG-------- --------((((..(((....(((((..........((((((((((......)))))))))).(((.((((....)))).))))))))....)))..)))).........-------- ( -26.20, z-score = -2.32, R) >consensus _________________CUCCCUUUGUACUUGUCAUUUAUGAAAAUUUGUUUAUUUUCGUAAUGCUCCAUUUGCAAAUGUAGCCAAAGUAAAUAUUGCAGCUUCGUUUG_________ .....................(((((..........((((((((((......)))))))))).(((.((((....)))).)))))))).............................. (-13.65 = -13.66 + 0.01)

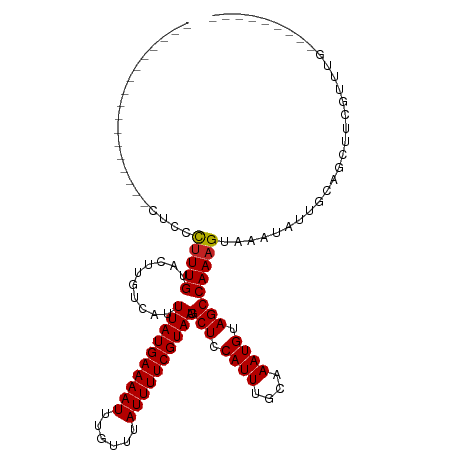
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:44 2011