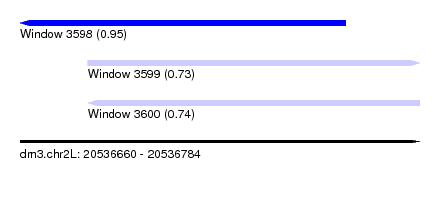
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,536,660 – 20,536,784 |
| Length | 124 |
| Max. P | 0.949384 |

| Location | 20,536,660 – 20,536,761 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.30 |
| Shannon entropy | 0.19448 |
| G+C content | 0.37937 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.45 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949384 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 20536660 101 - 23011544 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGA------------------- .................((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........))))))))...------------------- ( -25.93, z-score = -2.81, R) >dp4.Unknown_group_237 5732 108 + 11213 UCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGCACAAGGA------------ .....((..........((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))))).)).......------------ ( -26.23, z-score = -2.15, R) >droPer1.super_1 1045942 120 + 10282868 UCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGCAAAGGCACCAGCACAAGGA .....((..........((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))))).))....((....))....... ( -27.23, z-score = -1.06, R) >droAna3.scaffold_12916 7039142 102 - 16180835 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAAGGA------------------ .................((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........))))))))....------------------ ( -25.93, z-score = -2.80, R) >droEre2.scaffold_4845 18681413 113 + 22589142 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCACGAUUAUUUGUCUUCUUCAUUAGGCGAAACACGCAAAUCUCAAAGGCGAAGGCAAAGGAAAAGGA------- .....((..........(((.((((((((((....))))))))..)).)))...(((((((........)))))))....)).....((....((....))....))......------- ( -24.80, z-score = -1.23, R) >droYak2.chr2R 6892621 101 - 21139217 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGA------------------- .................((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........))))))))...------------------- ( -25.93, z-score = -2.81, R) >droSec1.super_18 419121 101 - 1342155 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGA------------------- .................((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........))))))))...------------------- ( -25.93, z-score = -2.81, R) >droSim1.chr2L 20107163 101 - 22036055 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGA------------------- .................((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........))))))))...------------------- ( -25.93, z-score = -2.81, R) >droVir3.scaffold_12963 6644395 95 + 20206255 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC------------------------- .................((((((((((((((....))))))))..))))))......(((((.........(((.....)))........)))))------------------------- ( -21.43, z-score = -1.97, R) >droMoj3.scaffold_6500 24614487 95 - 32352404 UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC------------------------- .................((((((((((((((....))))))))..))))))......(((((.........(((.....)))........)))))------------------------- ( -21.43, z-score = -1.97, R) >droGri2.scaffold_15126 6303024 95 - 8399593 UCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC------------------------- .................((((((((((((((....))))))))..))))))......(((((.........(((.....)))........)))))------------------------- ( -19.33, z-score = -1.07, R) >droWil1.scaffold_180708 10685286 108 + 12563649 UCACAGCAAUUAUUUACCUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGGAAAAGCA------------ ................(((((((((((((((....))))))))..)))).....((((((((.........(((.....)))........))))))))))).......------------ ( -25.53, z-score = -1.66, R) >consensus UCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGA___________________ .................((((((((((((((....))))))))..))))))......(((((.........(((.....)))........)))))......................... (-21.50 = -21.45 + -0.06)

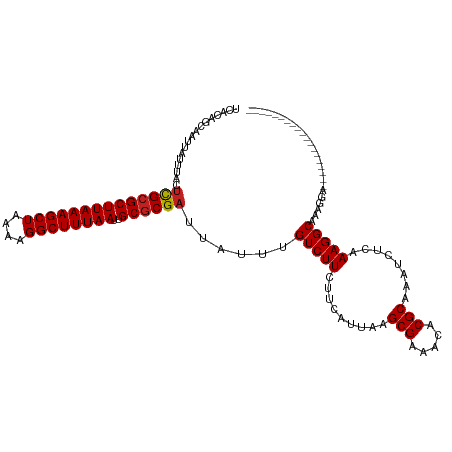

| Location | 20,536,681 – 20,536,784 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Shannon entropy | 0.15686 |
| G+C content | 0.38955 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -23.79 |
| Energy contribution | -23.15 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734748 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 20536681 103 + 23011544 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACGCACGCACA--- ((((.((((....................((((((..(((((((......)))))))))))))((((....)))).))))((((..........)))))))).--- ( -28.70, z-score = -1.46, R) >dp4.Unknown_group_237 5760 102 - 11213 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCAAGUAAAUAAUUGCUGUGAGUGCAGAA-CGAGAACACGCACA--- ((((((((((((...................((((..(((((((......))))))))))).(((((....))))).)))))).....-...)))))).....--- ( -24.10, z-score = -0.55, R) >droPer1.super_1 1045982 102 - 10282868 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCAAGUAAAUAAUUGCUGUGAGUGCAGAA-CGAGAACACGCACA--- ((((((((((((...................((((..(((((((......))))))))))).(((((....))))).)))))).....-...)))))).....--- ( -24.10, z-score = -0.55, R) >droAna3.scaffold_12916 7039164 106 + 16180835 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACACACGCAAAGUG ((.(((..(.....)..))).))......((((((..(((((((......)))))))))))))........((((((((.((......))...)))).)))).... ( -27.30, z-score = -1.29, R) >droYak2.chr2R 6892642 102 + 21139217 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAA-CGAACACACGCACA--- ((((((((((((....((...........((((((..(((((((......)))))))))))))...........)).)))))).....-.)))))).......--- ( -27.35, z-score = -1.53, R) >droSec1.super_18 419142 103 + 1342155 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACGCACGCACA--- ((((.((((....................((((((..(((((((......)))))))))))))((((....)))).))))((((..........)))))))).--- ( -28.70, z-score = -1.46, R) >droSim1.chr2L 20107184 103 + 22036055 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACGCACGCACA--- ((((.((((....................((((((..(((((((......)))))))))))))((((....)))).))))((((..........)))))))).--- ( -28.70, z-score = -1.46, R) >droVir3.scaffold_12963 6644410 101 - 20206255 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAAAAUGAGGCACACUCA----- ((((((((.....................((((((..(((((((......)))))))))))))........((((.......))))..)))))))).....----- ( -28.60, z-score = -2.03, R) >droMoj3.scaffold_6500 24614502 101 + 32352404 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAAAAUGAAGCACGUACA----- ((((((((.....................((((((..(((((((......)))))))))))))........((((.......))))..)))))))).....----- ( -29.00, z-score = -2.37, R) >droGri2.scaffold_15126 6303039 97 + 8399593 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCAAGUAAAUAAUUGCUGUGAGUGCAAAAUGAAACACA--------- (((((((((((.........((.......))((((..(((((((......)))))))))))))))......((((.......))))...))))))).--------- ( -26.00, z-score = -2.38, R) >droWil1.scaffold_180708 10685314 101 - 12563649 GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCAGGUAAAUAAUUGCUGUGAGUGCAAAAUGAAACACGAACA----- (((((((((((....))).............((((..(((((((......))))))).(((((.(((....)))))))).))))....)))))))).....----- ( -25.80, z-score = -1.84, R) >consensus GUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACACACGCACA___ ((((((((((((...................((((..(((((((......))))))))))).(((((....))))).)))))).......)))))).......... (-23.79 = -23.15 + -0.64)

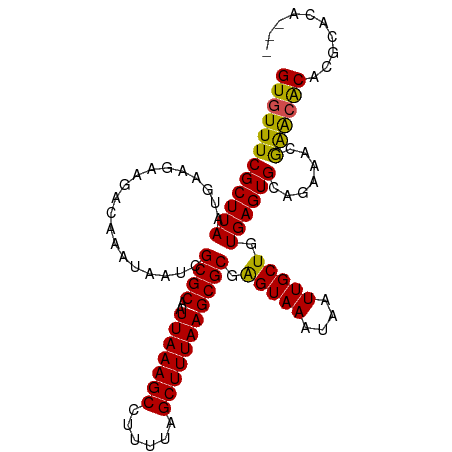
| Location | 20,536,681 – 20,536,784 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Shannon entropy | 0.15686 |
| G+C content | 0.38955 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -19.79 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743144 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 20536681 103 - 23011544 ---UGUGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---((((.(((((........))))).))))............((((((((((((((....))))))))..))))))...((((.(((.......))))))).... ( -28.80, z-score = -2.26, R) >dp4.Unknown_group_237 5760 102 + 11213 ---UGUGCGUGUUCUCG-UUCUGCACUCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---((((.((((.....-....)))).))))((............((((((((((((....))))))))..))))(((.....)))...........))....... ( -25.10, z-score = -1.57, R) >droPer1.super_1 1045982 102 + 10282868 ---UGUGCGUGUUCUCG-UUCUGCACUCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---((((.((((.....-....)))).))))((............((((((((((((....))))))))..))))(((.....)))...........))....... ( -25.10, z-score = -1.57, R) >droAna3.scaffold_12916 7039164 106 - 16180835 CACUUUGCGUGUGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ........((((.(((((((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))...........))))))))))) ( -27.80, z-score = -2.49, R) >droYak2.chr2R 6892642 102 - 21139217 ---UGUGCGUGUGUUCG-UUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---.....((((.((((-((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))............)))))))))) ( -27.00, z-score = -1.88, R) >droSec1.super_18 419142 103 - 1342155 ---UGUGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---((((.(((((........))))).))))............((((((((((((((....))))))))..))))))...((((.(((.......))))))).... ( -28.80, z-score = -2.26, R) >droSim1.chr2L 20107184 103 - 22036055 ---UGUGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---((((.(((((........))))).))))............((((((((((((((....))))))))..))))))...((((.(((.......))))))).... ( -28.80, z-score = -2.26, R) >droVir3.scaffold_12963 6644410 101 + 20206255 -----UGAGUGUGCCUCAUUUUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC -----((((((((........))))))))..............((((((((((((((....))))))))..))))))...((((.(((.......))))))).... ( -26.00, z-score = -2.28, R) >droMoj3.scaffold_6500 24614502 101 - 32352404 -----UGUACGUGCUUCAUUUUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC -----(((..((((........))))..)))............((((((((((((((....))))))))..))))))...((((.(((.......))))))).... ( -24.90, z-score = -2.39, R) >droGri2.scaffold_15126 6303039 97 - 8399593 ---------UGUGUUUCAUUUUGCACUCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC ---------.(((((((...((((.......)))).((..((.((((((((((((((....))))))))..)))))).))..))...............))))))) ( -25.80, z-score = -3.11, R) >droWil1.scaffold_180708 10685314 101 + 12563649 -----UGUUCGUGUUUCAUUUUGCACUCACAGCAAUUAUUUACCUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC -----.....(((((((...((((.......)))).........(((((((((((((....))))))))..))))).......................))))))) ( -24.90, z-score = -2.58, R) >consensus ___UGUGCGUGUGUUCGAUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACAC .........(((.........(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..)).............)))...... (-19.79 = -19.30 + -0.49)

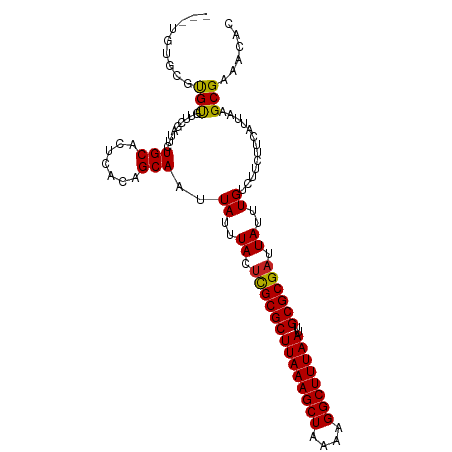
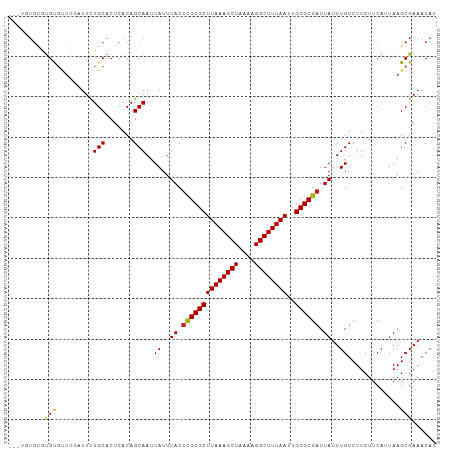
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:42 2011