
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,903,956 – 1,904,075 |
| Length | 119 |
| Max. P | 0.764609 |

| Location | 1,903,956 – 1,904,075 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.95 |
| Shannon entropy | 0.48725 |
| G+C content | 0.37719 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -12.16 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.764609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
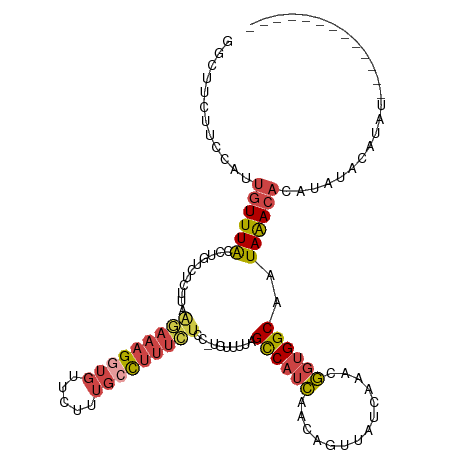
>dm3.chr2L 1903956 119 - 23011544 GGCUUCUUCCAUUGUUUACCUGUCUCUUAAGAAAGGUGUUCUUUGAUUUUCUCC-UGUUUUAGCCAUCAACAGUUAUCAAACGGUGGCAAUAAACAGAUAUACAUAUAUAUAUAUAUUUG .........((..(..(((((.(((....))).)))))..)..))........(-(((((..((((((..............))))))...))))))(((((......)))))....... ( -23.14, z-score = -1.71, R) >droAna3.scaffold_12916 495120 100 + 16180835 GGCCACUUGGCUUGUUUGCGAGGCU---AAAUAAGUUGUUUUCUCCCCAUU----GUUUCGGGCCAAGAACAGUUACCAAGACGGGGCAAUAAAAUGGUAAGUAAAU------------- .(((.(((((((((...((((((..---((((.....)))).....)).))----))..)))))))))........((.....)))))...................------------- ( -23.50, z-score = 0.51, R) >droEre2.scaffold_4929 1947614 103 - 26641161 GGCCUAUUCCAUUGUUUACCUGUCUC--AGAAAAGGUGUUCUUUGCCUUUCUCC-UGUUUUAGUCAUCAACAGUUAUCAAACGGUGGCAAUAAACAUAUAUAUUUG-------------- ....(((((((((((((.........--(((((.((((.....))))))))).(-((((.........))))).....))))))))).))))..............-------------- ( -18.30, z-score = -1.08, R) >droYak2.chr2L 1883381 105 - 22324452 GGCUUCUAUCAUUGUUUACCUGUCUCCCAAGAAAGGUGUUCUCUGCCUUUCUCC-GGUUUUAGCCAUCAACAGUUAUCAACCGGUGGCAAUAGACAUGUAUAUUUG-------------- ................(((.(((((.(((((((((((.......))))))))((-((((.((((........))))..)))))))))....))))).)))......-------------- ( -27.10, z-score = -2.74, R) >droSec1.super_14 1847564 117 - 2068291 GGCUUCUUCCAUUGUUUACCUGUCUCUUGAGAAAGGUGUUCUUUGCUUUUCUCC-UGUUUUAGCCAUCAACAGUUAUCAAACGGUGGCAAUAAACACAUAUACAUAUAAAUAUAUUUG-- ((......))..((((((..(((.....((((((((((.....)))))))))).-(((((..((((((..............))))))...))))).....)))..))))))......-- ( -23.34, z-score = -1.96, R) >droSim1.chr2L 1871330 118 - 22036055 GGCUUCUUCCAUUGUUUACCUGUCUCUUGAGAAAGGUGUUCUUGGCUUUUCUCCUUUUUUUAGCCAUCAACAGUUAUCAAACGGUGGCAAUAAACACAUAUUCAUAUAAAUAUAUUUG-- ........(((((((((((((.(((....))).))))(((..(((((..............)))))..))).......)))))))))...............................-- ( -23.44, z-score = -2.02, R) >consensus GGCUUCUUCCAUUGUUUACCUGUCUCUUAAGAAAGGUGUUCUUUGCCUUUCUCC_UGUUUUAGCCAUCAACAGUUAUCAAACGGUGGCAAUAAACACAUAUACAUAU_____________ ............((((((...........(((((((((.....)))))))))..........((((((..............))))))..))))))........................ (-12.16 = -13.06 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:38 2011