
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,513,334 – 20,513,425 |
| Length | 91 |
| Max. P | 0.691720 |

| Location | 20,513,334 – 20,513,425 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.38835 |
| G+C content | 0.39304 |
| Mean single sequence MFE | -16.74 |
| Consensus MFE | -10.44 |
| Energy contribution | -10.37 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20513334 91 - 23011544 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUACGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGAC-----CGGUCCA---UUCCACCU------------ ........(((((.(((....)))-..)))))......(((.(((((((....))))...((((.......))))..-----.))).))---).......------------ ( -15.70, z-score = -2.01, R) >droAna3.scaffold_12916 7010455 99 - 16180835 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGAC-----CCGGCCAGCGUGCCACCUCAUUU------- .....(((((((.(..((((((..-........))))))..).((((((....))))...((((.......))))..-----..)).)))))))...........------- ( -16.30, z-score = -0.80, R) >droEre2.scaffold_4845 18659760 91 + 22589142 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUACGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCACAC-----CGGACCA---UUCCACCU------------ ........(((((.(((....)))-..)))))......((((...((((....))))....(((....)))))))..-----.(((...---.)))....------------ ( -14.60, z-score = -2.32, R) >droYak2.chr2R 6868775 91 - 21139217 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUACGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCACAC-----CGGACCA---UUCCACCU------------ ........(((((.(((....)))-..)))))......((((...((((....))))....(((....)))))))..-----.(((...---.)))....------------ ( -14.60, z-score = -2.32, R) >droSec1.super_18 391402 91 - 1342155 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUACGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGAC-----CGGUCCA---UUCCACCU------------ ........(((((.(((....)))-..)))))......(((.(((((((....))))...((((.......))))..-----.))).))---).......------------ ( -15.70, z-score = -2.01, R) >droSim1.chr2L 20083537 91 - 22036055 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUACGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGAC-----CGGUCCA---UUCCACCU------------ ........(((((.(((....)))-..)))))......(((.(((((((....))))...((((.......))))..-----.))).))---).......------------ ( -15.70, z-score = -2.01, R) >droWil1.scaffold_180708 10645454 92 + 12563649 UAAUUCAUGGUGAUUGUAUGAACAUUUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACGUUUCAUCAACUAAAAAAAAAACAAAA-------------------- .......((((((.(((.((((....))))..(((((((.....)))))))............)))..))))))..................-------------------- ( -9.90, z-score = 0.38, R) >droPer1.super_1 1020819 90 + 10282868 UUAUUCAUGCUGAUUGUAUGAACA--UUCAGC-UUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCGGGC-----CCGGCCCG--GCCCCCCU------------ ...((((((((((.(((....)))--.)))))-.....)))))..((((....))))................((((-----(......)--))))....------------ ( -21.10, z-score = -2.04, R) >dp4.chr4_group3 3875641 92 + 11692001 UUAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCGGGC-----CCAGCCCG--GCCCACCU------------ ....(((.(((((.(((....)))-..)))))(((((((.....)))))))..........))).........((((-----(......)--))))....------------ ( -21.10, z-score = -2.49, R) >droVir3.scaffold_12963 6621180 107 + 20206255 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGGCAAC---AGCCCAGAACUCAAACCC-GAACUCAAGC ...(((..(((((.(((....)))-..)))))(((((((.....)))))))........(((((.......)))))....---..................-)))....... ( -15.20, z-score = -0.08, R) >droMoj3.scaffold_6500 24586167 108 - 32352404 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGCCAAC--AAGACCUGAGCUCCAACCC-GAGCUCAAGC ........(((((.(((....)))-..)))))(((((((.....)))))))(((......((((.......)))).....--)))...(((((((......-)))))))... ( -22.00, z-score = -2.18, R) >droGri2.scaffold_15126 6280165 111 - 8399593 UAAUUCAUGCUGAUUGUAUGAACA-UUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGGCAGCAGCAUCCCCGAACUUGAACCCAAAGCUCAACC ...(((((((.....)))))))..-.....(((((((((.....))))))......((.(((((.......))))).)).)))......((.(((.......))).)).... ( -19.00, z-score = -0.98, R) >consensus UAAUUCAUGCUGAUUGUAUGAACA_UUUCAGCUUUUAUGUGAACCGUAAAACUUUACUUCCUGACUUUUCAUCAGAC_____CGGACCAG__UUCCACCU____________ ....(((.(((((.(((....)))...)))))(((((((.....)))))))..........)))................................................ (-10.44 = -10.37 + -0.08)

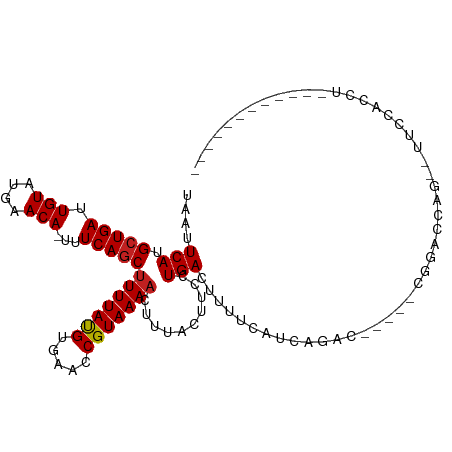
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:37 2011