| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,507,412 – 20,507,514 |
| Length | 102 |
| Max. P | 0.932507 |

| Location | 20,507,412 – 20,507,514 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 65.98 |
| Shannon entropy | 0.67318 |
| G+C content | 0.51463 |
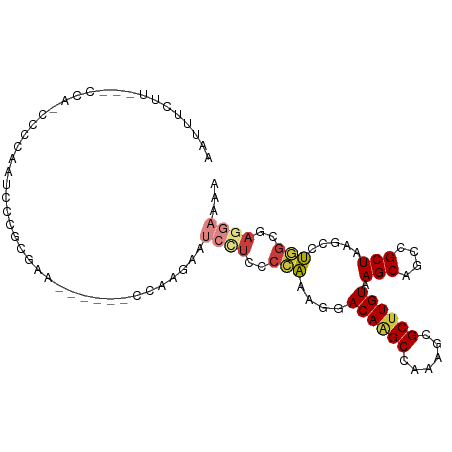
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -11.24 |
| Energy contribution | -11.93 |
| Covariance contribution | 0.69 |
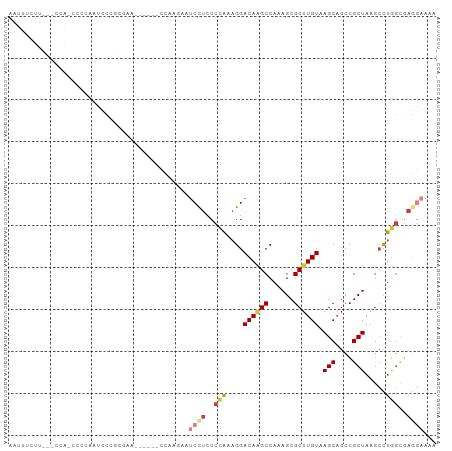
| Combinations/Pair | 1.25 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
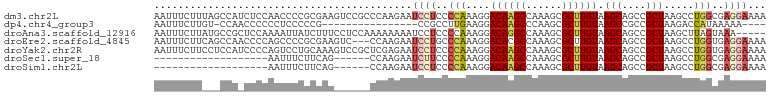
>dm3.chr2L 20507412 102 - 23011544 AAUUUCUUUAGCCAUCUCCAACCCCGCGAAGUCCGCCCAAGAAUCCUCCCCAAAGGACAAGCCAAAGCGCUUGUAAGCAGCCGCUAAGCCUGGCGAGGAAAA ...(((((..((...((.(........).))...))..)))))(((((.(((..((((((((......)))))).(((....)))...))))).)))))... ( -24.80, z-score = -1.02, R) >dp4.chr4_group3 3868091 81 + 11692001 AAUUUCUUGU-CCAACCCCCCUCCCCCG----------------CCGCCUUGAAGGACAAGCCCAAGCGCUUGUAAGCCGCCGCUAAGACCAUAAAAA---- .....(((((-((...............----------------..........))))))).....(((((....)).))).................---- ( -10.71, z-score = 0.23, R) >droAna3.scaffold_12916 7004347 97 - 16180835 AAUUUCUUAUGCCGCUCCAAAAUUAUCUUUCCUCCAAAAAAAAUCCUCCCCAAAGGACAGGCCAAAGCGCUUGUAAGCAGCCGCUAAGCUUAGUAAA----- .....((((.((.(((.(..........................(((......)))((((((......))))))..).))).)))))).........----- ( -15.70, z-score = -0.37, R) >droEre2.scaffold_4845 18653816 99 + 22589142 AAUUUCUUCAGCCAACCCCAGCCCCGCGAAGUC---CCAAGAAUCCUGCCCAAAGGACACGCCAAAGCGCUUGUAAGCAGCCGCUAAGCCUGGUGAGGAAAA ..((((((((.(((...........(((..(((---(.................)))).)))...((((.(((....))).)))).....))))))))))). ( -24.63, z-score = -0.77, R) >droYak2.chr2R 6862928 102 - 21139217 AAUUUCUUCCUCCAUCCCCAGUCCUGCAAAGUCCGCUCGAGAAUCCUCCCCAAAGGACAAGCCAAAGCGCUUGUAAGCAGCCGCUAAGCCUGGUGAGGAAAA ..(((((((..(((..(..(((.((((...((((....(((....)))......))))((((......))))....))))..)))..)..))).))))))). ( -27.00, z-score = -1.82, R) >droSec1.super_18 385535 77 - 1342155 -------------------AAUUUCUUCAG------CCAAGAAUCUUCCCCAAAGGACAAGCCAAAGCGCUUGUAAGCAGCCGCUAAGCCUGGCGAGGAAAA -------------------..(((((((.(------(((.......(((.....)))...((...((((.(((....))).))))..)).))))))))))). ( -21.90, z-score = -1.27, R) >droSim1.chr2L 20077702 77 - 22036055 -------------------AAUUUCUUCAG------CCAAGAAUCCUCCCCAAAGGACAAGCCAAAGCGCUUGUAAGCAGCCGCUAAGCCUGGCGAGGAAAA -------------------..(((((((.(------(((....((((......))))...((...((((.(((....))).))))..)).))))))))))). ( -23.50, z-score = -1.94, R) >consensus AAUUUCUU___CCA_CCCCAAUCCCGCGAA______CCAAGAAUCCUCCCCAAAGGACAAGCCAAAGCGCUUGUAAGCAGCCGCUAAGCCUGGCGAGGAAAA ...........................................((((..(((....((((((......)))))).(((....))).....)))..))))... (-11.24 = -11.93 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:35 2011