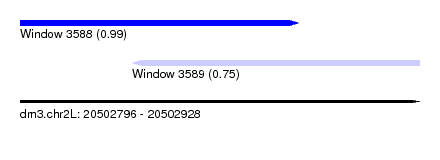
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,502,796 – 20,502,928 |
| Length | 132 |
| Max. P | 0.988994 |

| Location | 20,502,796 – 20,502,888 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.92 |
| Shannon entropy | 0.46167 |
| G+C content | 0.64587 |
| Mean single sequence MFE | -41.05 |
| Consensus MFE | -27.71 |
| Energy contribution | -29.80 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.988994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20502796 92 + 23011544 UCCGGGAGGCGUCCCUUGUGGUGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGC------------CACUCCCACUCGCAGCCCGGAGCAAC ((((((..(((......((((.((.(((.(((((...(((((((....))))))).)))))..)))------------...)))))).)))..))))))..... ( -44.70, z-score = -2.95, R) >droSim1.chr2L 20073120 104 + 22036055 UCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAAC ((((((..(((((((....)).)))(((.(((((...(((((((....))))))).)))))..))).......................))..))))))..... ( -43.80, z-score = -2.03, R) >droSec1.super_18 380878 104 + 1342155 UCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAAC ((((((..(((((((....)).)))(((.(((((...(((((((....))))))).)))))..))).......................))..))))))..... ( -43.80, z-score = -2.03, R) >droYak2.chr2R 6858298 104 + 21139217 CCCGGGAGGCAUCCCUUGUGGCAACGCUUGAUUGAAAUGCCUGGCAGGCCAGGCAGCGAUCCCAGCAACUCCCACUUCCACUCCCACUCGCCGCCCGGAGCAAC .(((((.(((.........(....)(((.(((((...(((((((....))))))).)))))..))).......................))).)))))...... ( -41.50, z-score = -2.31, R) >droEre2.scaffold_4845 18649687 90 - 22589142 -GCCGGGGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGCAGGCCAGGCAGCGAUCC-GGC------------CACGCCCACUCGCAGCCCGGGGCAAC -(((..((((......((.((((..(((.(((((...(((((((....))))))).))))).-)))------------..))))))......))))..)))... ( -46.30, z-score = -1.84, R) >droAna3.scaffold_12916 7000249 75 + 16180835 ACUGGAGCUCGUCUCUUGAGGCGGUGCUUGAUUGAAAUGCCUGGUAGGCAUUGCCUGGCUUU----------------CACCCCGAGCUAC------------- .....((((((.......((((((((((((...(......)...))))))))))))((....----------------..)).))))))..------------- ( -26.20, z-score = -1.33, R) >consensus UCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGC____________CACUCCCACUCGCAGCCCGGAGCAAC .((((((((((((((....)).)))))))(((((...(((((((....))))))).)))))................................)))))...... (-27.71 = -29.80 + 2.09)

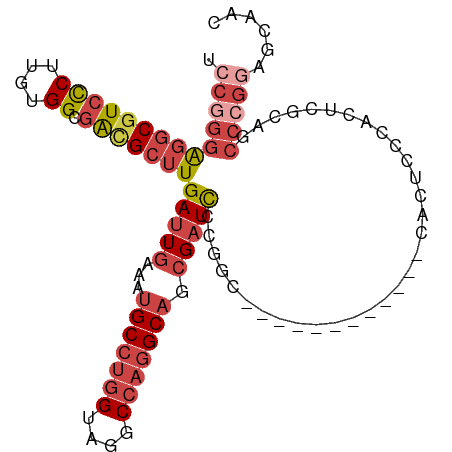
| Location | 20,502,833 – 20,502,928 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Shannon entropy | 0.34058 |
| G+C content | 0.57816 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -30.91 |
| Energy contribution | -33.63 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20502833 95 - 23011544 AAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCG------------AGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCA ............(((.((((.(((.((((((((((....)))))))))).)))..)------------)))((....(((((((.......)))))))..)).))). ( -46.90, z-score = -3.05, R) >droSim1.chr2L 20073157 106 - 22036055 -AAAAUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCA -...........(((.((((.(((.((((((((((....)))))))))).)))..))))...........(((....(((((((.......)))))))..)))))). ( -47.80, z-score = -2.39, R) >droSec1.super_18 380915 107 - 1342155 AAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCA ............(((.((((.(((.((((((((((....)))))))))).)))..))))...........(((....(((((((.......)))))))..)))))). ( -47.80, z-score = -2.06, R) >droYak2.chr2R 6858335 107 - 21139217 AAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGCCAGUUGCUCCGGGCGGCGAGUGGGAGUGGAAGUGGGAGUUGCUGGGAUCGCUGCCUGGCCUGCCAGGCA .............((.((((..((.((((((((((....)))))))))).))...)))).))(((((.((..(...)..))....)))))((((((....)))))). ( -43.10, z-score = -0.46, R) >droEre2.scaffold_4845 18649723 94 + 22589142 AAAACUAAUUAAGCCAACUUAAGUGCGGACCAGCUUGCCAGUUGCCCCGGGCUGCG------------AGUGGGCGUGGCC-GGAUCGCUGCCUGGCCUGCCAGGCA ............(((.((((.(((.(((..(((((....)))))..))).)))..)------------))).((((.((((-((........)))))))))).))). ( -39.60, z-score = -0.54, R) >droAna3.scaffold_12916 7000286 78 - 16180835 AAAACUAAUUAAGCCAACUUAAGUGCGGAGCAUCUUGGCAGUAGCUCGGG-------------------GUGAAA---GCCAG-------GCAAUGCCUACCAGGCA ............(((((.....((((...)))).)))))....((..(((-------------------((....---)))((-------((...)))).))..)). ( -22.60, z-score = -0.09, R) >consensus AAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCG____________AGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCA ............(((......(((.((((((((((....)))))))))).))).................(((....(((((((.......)))))))..)))))). (-30.91 = -33.63 + 2.72)
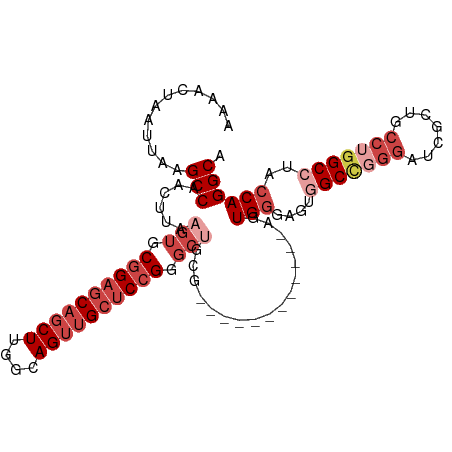
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:33 2011