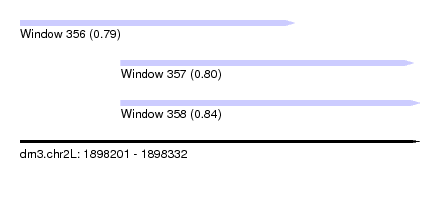
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,898,201 – 1,898,332 |
| Length | 131 |
| Max. P | 0.841104 |

| Location | 1,898,201 – 1,898,291 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Shannon entropy | 0.48634 |
| G+C content | 0.43262 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -13.49 |
| Energy contribution | -12.99 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786542 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
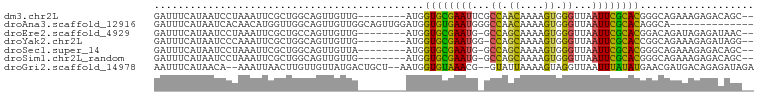
>dm3.chr2L 1898201 90 + 23011544 GAUUUCAUAAUCCUAAAUUCGCUGGCAGUUGUUG--------AUGGUGCGAAUUCGCCAACAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGC-- ((((....))))........((((.......(((--------.(.(((((((((.(((.((....)).))).))))))))).).))).......))))-- ( -26.04, z-score = -1.89, R) >droAna3.scaffold_12916 489838 86 - 16180835 GAUUUCAUAAUCACAACAUGGUUGGCAGUUGUUGGCAGUUGGAUGGUGUGAAUGGGCCAACAAAAGUGGGUUAAUUCGCACAGGCA-------------- .........(((.((((...((..((....))..)).))))))).((((((((..(((.((....)).)))..)))))))).....-------------- ( -27.60, z-score = -2.40, R) >droEre2.scaffold_4929 1941837 89 + 26641161 GAUUUCAUAAUCCUAAAUUCGCUGCCAGUUGUUG--------AUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGACAGAUAGAGAUAAC-- .(((((((..(((...(((.((........)).)--------)).(((((((((-(((.((....)).)))).)))))))))))...)).)))))...-- ( -22.50, z-score = -1.08, R) >droYak2.chr2L 1877550 89 + 22324452 GAUUUCAUAAUCCCAAAUUCGCUGGCAGUUGUUG--------AUGGUGCGAAUGG-CCAGCAAAAGUGGGUUAAUUCGCACCGGCAGAAAGAGAUAGG-- .(((((.(((.((((..((.((((((..((((..--------.....))))...)-))))).))..))))))).(((((....)).))).)))))...-- ( -26.80, z-score = -1.72, R) >droSec1.super_14 1841802 89 + 2068291 GAUUUCAUAAUCCUAAAUUCGCUGGCAGUUGUUA--------AUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGC-- ((((....))))........((((.(..(((((.--------...(((((((((-(((.((....)).)))).)))))))).)))))...)...))))-- ( -24.00, z-score = -1.20, R) >droSim1.chr2L_random 106023 89 + 909653 GAUUUCAUAAUCCUAAAUUCGCUGGCAGUUGUUG--------AUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGC-- ((((....))))........((((.......(((--------.(.(((((((((-(((.((....)).)))).)))))))).).))).......))))-- ( -24.84, z-score = -1.15, R) >droGri2.scaffold_14978 939834 94 + 1124632 AAUUUCAUAACA--AAAUUAACUUGUUGUUAUGACUGCU--AAUGGUGUAAACG--GUAUUAAAAGUAGGUUAAUUUAUAUGAACGAUGACAGAGAUAGA ...((((((...--((((((((((.........(((..(--((((.((....))--.)))))..))))))))))))).))))))................ ( -14.10, z-score = -0.19, R) >consensus GAUUUCAUAAUCCUAAAUUCGCUGGCAGUUGUUG________AUGGUGCGAAUG_GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGC__ .............................................((((((((..(((.((....)).)))..))))))))................... (-13.49 = -12.99 + -0.50)
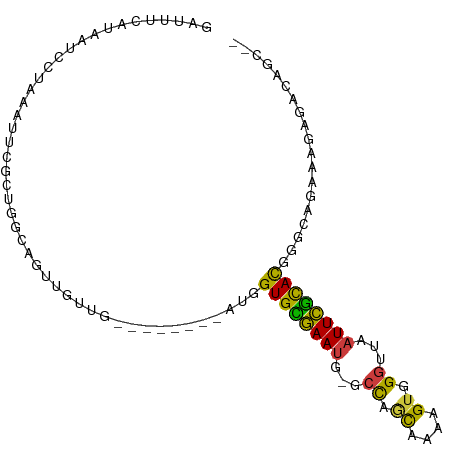
| Location | 1,898,234 – 1,898,330 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Shannon entropy | 0.13015 |
| G+C content | 0.52509 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1898234 96 + 23011544 GAUGGUGCGAAUUCGCCAACAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCAGAUG-GUCGGGCGAAAGAGAUGGCAACAGAUGUGAAUAG ....(((((((((.(((.((....)).))).)))))))))...............(((..((-((((..(....)...))))..))..)))...... ( -26.50, z-score = -2.57, R) >droEre2.scaffold_4929 1941870 95 + 26641161 GAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGACAGAUAGAGAUAACCGAUG-GGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAG ....(((((((((-(((.((....)).)))).))))))))...((.((........(((...-..)))..........((....)).)).))..... ( -23.40, z-score = -1.94, R) >droYak2.chr2L 1877583 95 + 22324452 GAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACCGGCAGAAAGAGAUAGGUGAUG-GGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAG ..(((((((((((-(((.((....)).)))).))))))))))..........(((..((...-(.((..(....)...)).)..))..)))...... ( -25.00, z-score = -2.39, R) >droSec1.super_14 1841835 96 + 2068291 AAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCCGAAGAGGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAG ....(((((((((-(((.((....)).)))).))))))))..(((......(...(((.....)))...)........((....))..)))...... ( -25.60, z-score = -2.29, R) >droSim1.chr2L_random 106056 96 + 909653 GAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCCGACGAGGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAG ....(((((((((-(((.((....)).)))).))))))))..(((......(...(((.....)))...)........((....))..)))...... ( -25.60, z-score = -1.87, R) >consensus GAUGGUGCGAAUG_GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCCGAUG_GGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAG ....((((((((..(((.((....)).)))..))))))))..(((...........(((......)))..........((....))..)))...... (-20.04 = -20.00 + -0.04)

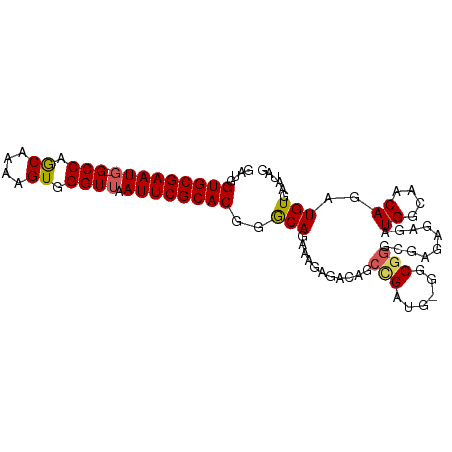
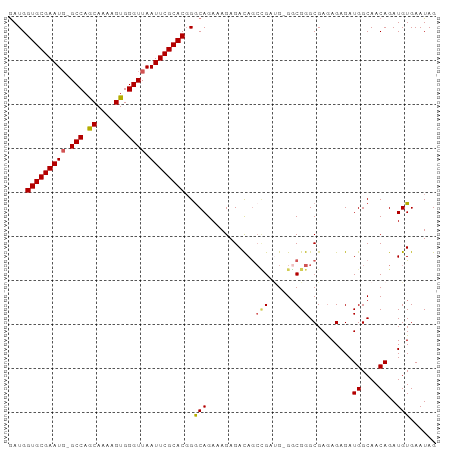
| Location | 1,898,234 – 1,898,332 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Shannon entropy | 0.31642 |
| G+C content | 0.51122 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -18.45 |
| Energy contribution | -18.29 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1898234 98 + 23011544 -----GAUGGUGCGAAUUCGCCAACAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCAGAUG-GUCGGGCGAAAGAGAUGGCAACAGAUGUGAAUAGCU -----....(((((((((.(((.((....)).))).)))))))))...............(((..((-((((..(....)...))))..))..)))........ ( -26.50, z-score = -1.88, R) >droAna3.scaffold_12916 489874 90 - 16180835 AGUUGGAUGGUGUGAAUGGGCCAACAAAAGUGGGUUAAUUCGCACAGGCAAGUGG--------------CUAAGAGAGAGAAAUAGAGACAGAUGUGAAUAGCU (((((....((((((((..(((.((....)).)))..)))))))).(((.....)--------------))............................))))) ( -19.90, z-score = -1.97, R) >droEre2.scaffold_4929 1941870 97 + 26641161 -----GAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGACAGAUAGAGAUAACCGAUG-GGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAGCU -----....(((((((((-(((.((....)).)))).))))))))................(((...-..)))((........((....))..........)). ( -23.57, z-score = -1.26, R) >droYak2.chr2L 1877583 97 + 22324452 -----GAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACCGGCAGAAAGAGAUAGGUGAUG-GGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAGCU -----...((((((((((-(((.((....)).)))).)))))))))(((..................-.((.(.(....)...).)).((....)).....))) ( -25.30, z-score = -1.59, R) >droSec1.super_14 1841835 98 + 2068291 -----AAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCCGAAGAGGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAGCU -----....(((((((((-(((.((....)).)))).)))))))).(((.......(...(((.....)))...)...(.(..((....))..).).....))) ( -27.20, z-score = -2.00, R) >droSim1.chr2L_random 106056 98 + 909653 -----GAUGGUGCGAAUG-GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCCGACGAGGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAGCU -----....(((((((((-(((.((....)).)))).)))))))).(((.......(...(((.....)))...)...(.(..((....))..).).....))) ( -27.20, z-score = -1.67, R) >consensus _____GAUGGUGCGAAUG_GCCAGCAAAAGUGGGUUAAUUCGCACGGGCAGAAAGAGACAGCCGAUG_GGCGGGCGAGAGAGAUGGCAACAGAUGUGAAUAGCU .........((((((((..(((.((....)).)))..))))))))...........................(((........((....))..........))) (-18.45 = -18.29 + -0.16)

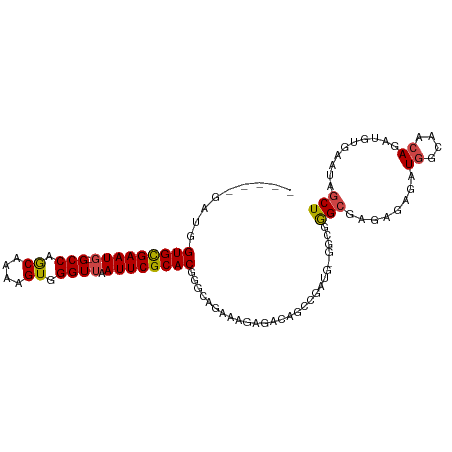
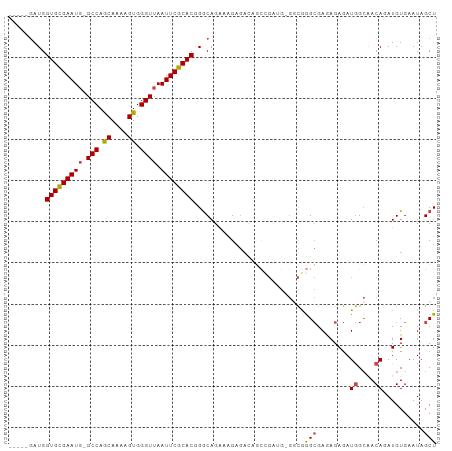
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:37 2011