| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,487,432 – 20,487,536 |
| Length | 104 |
| Max. P | 0.999573 |

| Location | 20,487,432 – 20,487,536 |
|---|---|
| Length | 104 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Shannon entropy | 0.40770 |
| G+C content | 0.40873 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -29.61 |
| Energy contribution | -29.58 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -5.08 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
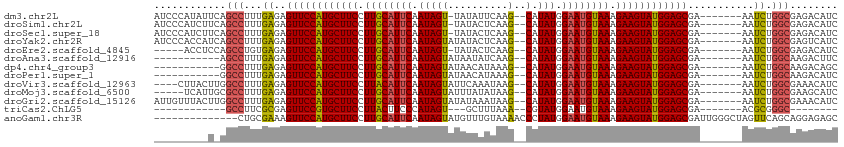
>dm3.chr2L 20487432 104 + 23011544 AUCCCAUAUUCAGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGU-UAUAUUCAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAGACAUC ........(((.(((...((..((((((((((((.((((((((.(((((-(.......))--).))).)))))))).))))))))))))..-------..)).))))))..... ( -36.50, z-score = -4.93, R) >droSim1.chr2L 20052700 104 + 22036055 AUCCCAUCUUCAGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGU-UAUACUCAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAGACAUC ......((((..(((...((..((((((((((((.((((((((.(((((-(.......))--).))).)))))))).))))))))))))..-------..)).))))))).... ( -37.10, z-score = -4.76, R) >droSec1.super_18 365657 104 + 1342155 AUCCCAUCUUCAGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGU-UAUACUCAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAGACAUC ......((((..(((...((..((((((((((((.((((((((.(((((-(.......))--).))).)))))))).))))))))))))..-------..)).))))))).... ( -37.10, z-score = -4.76, R) >droYak2.chr2R 6842715 105 + 21139217 AUCCCACCAUCAGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUAUACUCAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAGUCAUC ............(((...((..((((((((((((.((((((((.(((((..........)--).))).)))))))).))))))))))))..-------..)).)))........ ( -35.00, z-score = -4.32, R) >droEre2.scaffold_4845 18634774 99 - 22589142 -----ACCUCCAGCCUGUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGU-UAUACUCAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAGACAUC -----..(((..(((...((..((((((((((((.((((((((.(((((-(.......))--).))).)))))))).))))))))))))..-------..)).))))))..... ( -37.40, z-score = -5.02, R) >droAna3.scaffold_12916 6985824 94 + 16180835 -----------AGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUAAUAUCAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCAAGACUUC -----------.(((...((..((((((((((((.((((((((.(((((..........)--).))).)))))))).))))))))))))..-------..)).)))........ ( -35.00, z-score = -5.34, R) >dp4.chr4_group3 3846590 94 - 11692001 -----------GGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUAACAUAAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCAAGACAGC -----------.(((...((..((((((((((((.((((((((.(((((..........)--).))).)))))))).))))))))))))..-------..)).)))........ ( -34.80, z-score = -5.27, R) >droPer1.super_1 990758 94 - 10282868 -----------GGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUAACAUAAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCAAGACAUC -----------.(((...((..((((((((((((.((((((((.(((((..........)--).))).)))))))).))))))))))))..-------..)).)))........ ( -34.80, z-score = -5.36, R) >droVir3.scaffold_12963 6592732 101 - 20206255 ----CUUACUUGGCCUUUGAGAGUUCCAUGCUUCCUUACAUUCAAUAGUAUUCAAAUAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAAACAUC ----.....((.(((...((..((((((((((((.((((((((....((((.(......)--.)))).)))))))).))))))))))))..-------..)).))).))..... ( -36.30, z-score = -5.67, R) >droMoj3.scaffold_6500 24555181 100 + 32352404 -----UCAUUGCGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUUUAUAUAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAAGCAUC -----......((((...((..((((((((((((.((((((((.(((.(((....)))..--..))).)))))))).))))))))))))..-------..)).))))....... ( -37.10, z-score = -5.07, R) >droGri2.scaffold_15126 6250403 105 + 8399593 AUUGUUUACUUGGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUAUAAAUAAG--CAUAUGGAAUGUAAAGAAGUAUGGAGCGA-------AAUCUGGCGAAACAUC ..(((((.....(((...((..((((((((((((.((((((((.(((((.((....)).)--).))).)))))))).))))))))))))..-------..)).))).))))).. ( -36.20, z-score = -5.12, R) >triCas2.ChLG5 7115717 82 - 18847211 ------------GCCUUCGCGAGUUCCGUGCUUCCUUACUUCCCAUAGU---GCUUUAAA--CGUAUGGAAUGUAAAGAAGUAUGGAGCGA-------ACGCGGGC-------- ------------(((..((((.((((((((((((.((((...(((((((---.......)--).)))))...)))).))))))))))))..-------.)))))))-------- ( -36.80, z-score = -5.50, R) >anoGam1.chr3R 42702883 100 - 53272125 --------------CUGCGAAAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUGUUUGUAAAACCCUAUGGAAUGUAAAGAAGUAUGGAGCGAUUGGGCUAGUUCAGCAGGAGAGC --------------((((....((((((((((((.((((((((.((((...(((.....))).)))).)))))))).))))))))))))...((((....))))))))...... ( -37.80, z-score = -4.94, R) >consensus _________U_AGCCUUUGAGAGUUCCAUGCUUCCUUGCAUUCAAUAGUAUAUAAUCAAG__CAUAUGGAAUGUAAAGAAGUAUGGAGCGA_______AAUCUGGCGAGACAUC ............(((...((..((((((((((((.((((((((.(((.................))).)))))))).))))))))))))...........)).)))........ (-29.61 = -29.58 + -0.03)

| Location | 20,487,432 – 20,487,536 |
|---|---|
| Length | 104 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Shannon entropy | 0.40770 |
| G+C content | 0.40873 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
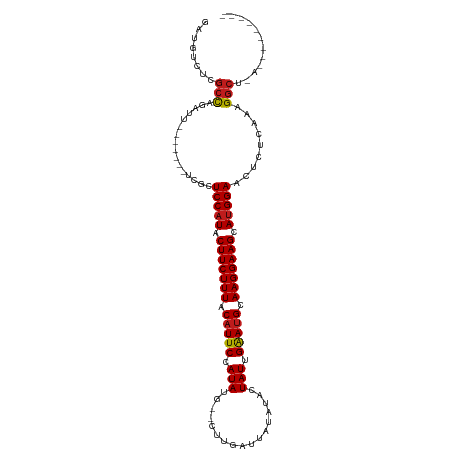
>dm3.chr2L 20487432 104 - 23011544 GAUGUCUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUGAAUAUA-ACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCUGAAUAUGGGAU ......((.(((.(((-------(.(((((((.(((((((.(((((.(((..--..........-..))).))))).))))))).))))).((.....)))).)))).))))). ( -23.84, z-score = -1.00, R) >droSim1.chr2L 20052700 104 - 22036055 GAUGUCUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUGAGUAUA-ACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCUGAAGAUGGGAU ......((.(((..((-------(.(((((((.(((((((.(((((..((((--(....)))))-......))))).))))))).))))).((.....)))).)))..))))). ( -26.40, z-score = -1.12, R) >droSec1.super_18 365657 104 - 1342155 GAUGUCUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUGAGUAUA-ACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCUGAAGAUGGGAU ......((.(((..((-------(.(((((((.(((((((.(((((..((((--(....)))))-......))))).))))))).))))).((.....)))).)))..))))). ( -26.40, z-score = -1.12, R) >droYak2.chr2R 6842715 105 - 21139217 GAUGACUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUGAGUAUAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCUGAUGGUGGGAU .....(((((((....-------....(((((.(((((((.(((((.(((((--(....))))))......))))).))))))).)))))....(((.....)))))))))).. ( -30.40, z-score = -2.20, R) >droEre2.scaffold_4845 18634774 99 + 22589142 GAUGUCUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUGAGUAUA-ACUAUUGAAUGCAAGGAAGCAUGGAACUCUCACAGGCUGGAGGU----- .((.(((.(((.....-------....(((((.(((((((.(((((..((((--(....)))))-......))))).))))))).))))).........))).))).))----- ( -27.17, z-score = -1.56, R) >droAna3.scaffold_12916 6985824 94 - 16180835 GAAGUCUUGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUGAUAUUAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCU----------- (((((((....)))))-------))..(((((.(((((((.(((((.(((..--.............))).))))).))))))).))))).............----------- ( -23.26, z-score = -2.19, R) >dp4.chr4_group3 3846590 94 + 11692001 GCUGUCUUGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUUAUGUUAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCC----------- ...(((((...(((..-------..(.(((((.(((((((.(((((.(((.(--(.....)).....))).))))).))))))).))))).))))..))))).----------- ( -22.20, z-score = -1.87, R) >droPer1.super_1 990758 94 + 10282868 GAUGUCUUGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUUAUGUUAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCC----------- ...(((((...(((..-------..(.(((((.(((((((.(((((.(((.(--(.....)).....))).))))).))))))).))))).))))..))))).----------- ( -22.20, z-score = -1.90, R) >droVir3.scaffold_12963 6592732 101 + 20206255 GAUGUUUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUAUUUGAAUACUAUUGAAUGUAAGGAAGCAUGGAACUCUCAAAGGCCAAGUAAG---- ........(((.....-------....(((((.(((((((((((((.(((..--.............))).))))))))))))).))))).........)))........---- ( -25.43, z-score = -2.75, R) >droMoj3.scaffold_6500 24555181 100 - 32352404 GAUGCUUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUAUAUAAAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCGCAAUGA----- .......((((.....-------....(((((.(((((((.(((((.(((..--.............))).))))).))))))).))))).........))))......----- ( -22.63, z-score = -1.60, R) >droGri2.scaffold_15126 6250403 105 - 8399593 GAUGUUUCGCCAGAUU-------UCGCUCCAUACUUCUUUACAUUCCAUAUG--CUUAUUUAUAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCCAAGUAAACAAU ..(((((((((.....-------....(((((.(((((((.(((((.(((.(--..((....))..)))).))))).))))))).))))).........)))...).))))).. ( -21.97, z-score = -1.95, R) >triCas2.ChLG5 7115717 82 + 18847211 --------GCCCGCGU-------UCGCUCCAUACUUCUUUACAUUCCAUACG--UUUAAAGC---ACUAUGGGAAGUAAGGAAGCACGGAACUCGCGAAGGC------------ --------....((.(-------(((((((...(((((((((.(((((((.(--(.......---))))))))).)))))))))...)))....))))).))------------ ( -30.10, z-score = -4.18, R) >anoGam1.chr3R 42702883 100 + 53272125 GCUCUCCUGCUGAACUAGCCCAAUCGCUCCAUACUUCUUUACAUUCCAUAGGGUUUUACAAACAUACUAUUGAAUGCAAGGAAGCAUGGAACUUUCGCAG-------------- ......((((.(((...((......))(((((.(((((((.(((((.((((.(((.....)))...)))).))))).))))))).)))))...)))))))-------------- ( -29.00, z-score = -4.39, R) >consensus GAUGUCUCGCCAGAUU_______UCGCUCCAUACUUCUUUACAUUCCAUAUG__CUUGAUUAUAUACUAUUGAAUGCAAGGAAGCAUGGAACUCUCAAAGGCU_A_________ ........(((.((.........))..(((((.(((((((.(((((.(((.................))).))))).))))))).))))).........)))............ (-18.09 = -18.41 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:29 2011