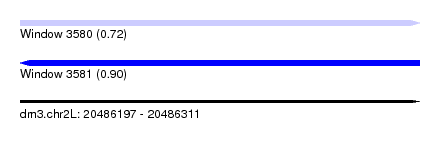
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,486,197 – 20,486,311 |
| Length | 114 |
| Max. P | 0.900365 |

| Location | 20,486,197 – 20,486,311 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Shannon entropy | 0.23548 |
| G+C content | 0.63376 |
| Mean single sequence MFE | -45.74 |
| Consensus MFE | -34.88 |
| Energy contribution | -34.56 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20486197 114 + 23011544 CGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGUCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGACCAUGCGACCAUGUGACCAUGUGCCCGCCCGACGCC .((((((....(((((.(....)...)))))(((((((((.(((.....)))))))))))).....)))))).((((.....(((..(((((....)))))..)))....)))) ( -44.50, z-score = -1.48, R) >droEre2.scaffold_4845 18633615 92 - 22589142 CGGUACACUUAGGGCGUCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAGCUGCAGAUUGCAUCCAUGUGCCAGGCGACCAUGUG-CCACGUC--------------------- .((((((.....((((.(((((.(......((((((((((.(((.....)))))))))))))......).))))))).)).))))-)).....--------------------- ( -41.30, z-score = -2.31, R) >droYak2.chr2R 6841528 113 + 21139217 CGGUACACUUAAGGCGUCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGACCAAGCGACCAUGUUACCAUGUGCC-GCCCGACGCC .((((((....((((((........))))))(((((((((.(........).))))))))).....)))))).((((.....(((.(((((....)))).).)-))....)))) ( -44.90, z-score = -2.35, R) >droSec1.super_18 364460 114 + 1342155 CGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGGCCAGGCGACCAUGCGACCAUAUGCCCGCCCGACGCC .((((((....(((((.(....)...)))))(((((((((.(........).))))))))).....)))))).((((..(.((((..((((......))))..)))).).)))) ( -48.30, z-score = -1.62, R) >droSim1.chr2L 20049675 114 + 22036055 CGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGGCCAGGCGACCAGGCGACCAUGUGCCCGCCCGACGCC .((((((....(((((.(....)...)))))(((((((((.(........).))))))))).....)))))).((((..(.((((....((((......)))))))).).)))) ( -49.70, z-score = -1.23, R) >consensus CGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGACCAGGCGACCAUGUGACCAUGUGCCCGCCCGACGCC .((((((....(((((.(....)...)))))(((((((((.(........).))))))))).....)))))).((((......)).)).......................... (-34.88 = -34.56 + -0.32)

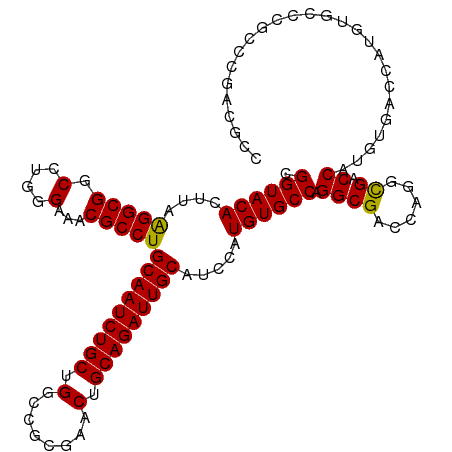
| Location | 20,486,197 – 20,486,311 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Shannon entropy | 0.23548 |
| G+C content | 0.63376 |
| Mean single sequence MFE | -50.84 |
| Consensus MFE | -39.51 |
| Energy contribution | -39.54 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20486197 114 - 23011544 GGCGUCGGGCGGGCACAUGGUCACAUGGUCGCAUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGACCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCG ((((((((((((.((..((..(....)..))..)).))))))))).......((((((((((.(((...)))..))))))))))(((.(.....).))))))............ ( -47.00, z-score = -0.89, R) >droEre2.scaffold_4845 18633615 92 + 22589142 ---------------------GACGUGG-CACAUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGCUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGACGCCCUAAGUGUACCG ---------------------..((..(-(((..((.(((((((...(((..((((((((((.((.....))..))))))))))..)))...))))).)).))...))))..)) ( -39.60, z-score = -1.92, R) >droYak2.chr2R 6841528 113 - 21139217 GGCGUCGGGC-GGCACAUGGUAACAUGGUCGCUUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGACGCCUUAAGUGUACCG ((((((((((-(((.((((....))))))))))))).)))).((.((((...((((((((((.((.....))..))))))))))((((((......))))))....)))).)). ( -56.20, z-score = -3.94, R) >droSec1.super_18 364460 114 - 1342155 GGCGUCGGGCGGGCAUAUGGUCGCAUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCG ((((((((((((.(((........))).))))))))).((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))............ ( -53.30, z-score = -1.36, R) >droSim1.chr2L 20049675 114 - 22036055 GGCGUCGGGCGGGCACAUGGUCGCCUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCG ((((((((((((.(....).)))))))).)))).(((.((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))............ ( -58.10, z-score = -2.32, R) >consensus GGCGUCGGGCGGGCACAUGGUCACAUGGUCGCAUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCG ...(((.....)))............((((((..(((.((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))....))).))). (-39.51 = -39.54 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:26 2011