| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,478,781 – 20,478,871 |
| Length | 90 |
| Max. P | 0.995469 |

| Location | 20,478,781 – 20,478,871 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 79.31 |
| Shannon entropy | 0.40166 |
| G+C content | 0.53244 |
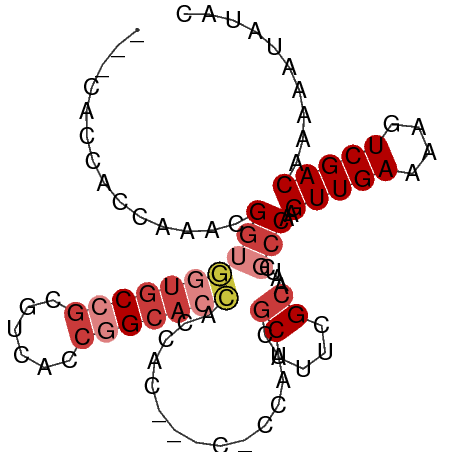
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -14.19 |
| Energy contribution | -15.70 |
| Covariance contribution | 1.51 |
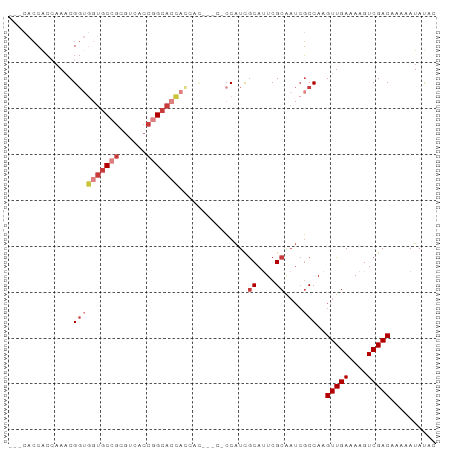
| Combinations/Pair | 1.06 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
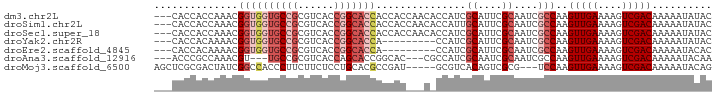
>dm3.chr2L 20478781 90 + 23011544 ---CACCACCAAACGGUGGUGCCGCGUCACCGGCACCACCACCAACACCAUCGCAUUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUAUAC ---...........((((((((((......))))))))))............((....)).........(((((....))))).......... ( -26.80, z-score = -3.77, R) >droSim1.chr2L 20042210 90 + 22036055 ---CACCACCAAACGGUGGUGCCGCGUCACCGGCACCACCACCAACACCAUUGCAUUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUAUAC ---...........((((((((((......)))))))))).........(((((....)))))......(((((....))))).......... ( -28.80, z-score = -4.22, R) >droSec1.super_18 356991 90 + 1342155 ---CACCACCAAACGGUGGUGCCGCGUCACCGGCACCACCACCAACACCAUCGCAUUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUAUAC ---...........((((((((((......))))))))))............((....)).........(((((....))))).......... ( -26.80, z-score = -3.77, R) >droYak2.chr2R 6834336 81 + 21139217 ---CACCACAAAACGGUGGUGCCGCGUCACCGGCACCA---------CCAUCGCAUUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUAUAC ---...........((((((((((......))))))))---------))...((....)).........(((((....))))).......... ( -26.80, z-score = -4.03, R) >droEre2.scaffold_4845 18626343 81 - 22589142 ---CACCACAAAACGGUGGUGCCGCGUCACCGGCACCA---------CCAUCGCAUUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUACAC ---...........((((((((((......))))))))---------))...((....)).........(((((....))))).......... ( -26.80, z-score = -3.89, R) >droAna3.scaffold_12916 6977797 84 + 16180835 ---ACCCGCCAAACGU---UGCCGCGUCACCAGCACCGGCAC---CGCCAUCGCAAUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUACAA ---....((....((.---(((((.(((....).))))))).---)).....((....))....))...(((((....))))).......... ( -15.80, z-score = -0.08, R) >droMoj3.scaffold_6500 24536296 85 + 32352404 AGCUCGCGACUAUCGGCCACCCUUCUUCUCCUGCACGCCGAU-----GCGUCACAGUCGCG---UCCAAGUUGAAAAGUCGACAAAAAUACAG .(..(((((((((((((...................))))))-----.......)))))))---..)..(((((....))))).......... ( -21.22, z-score = -1.44, R) >consensus ___CACCACCAAACGGUGGUGCCGCGUCACCGGCACCACCAC___C_CCAUCGCAUUCGCAAUCGCCAAGUUGAAAAGUCGACAAAAAUAUAC ..............((((((((((......)))))))...............((....))....)))..(((((....))))).......... (-14.19 = -15.70 + 1.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:24 2011