
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,452,280 – 20,452,387 |
| Length | 107 |
| Max. P | 0.821985 |

| Location | 20,452,280 – 20,452,387 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.40760 |
| G+C content | 0.65033 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -29.80 |
| Energy contribution | -30.83 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
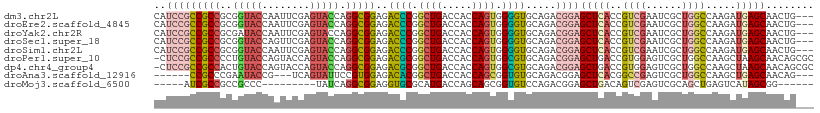
>dm3.chr2L 20452280 107 + 23011544 CAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGACGGAGCUCACCGUCGAAUCGCUGGCCAAGAUGAGCAACUG--- ..(((((((((..(((((........))))).)))))..((((.((((.....)))).)))).....))))((((((.((((......))))...).))))).....--- ( -43.60, z-score = -1.73, R) >droEre2.scaffold_4845 18599672 107 - 22589142 CAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGACGGAGCUCACCGUCGAAUCGCUGGCCAAGAUGAGCAACUG--- ..(((((((((..(((((........))))).)))))..((((.((((.....)))).)))).....))))((((((.((((......))))...).))))).....--- ( -43.60, z-score = -1.73, R) >droYak2.chr2R 6808490 107 + 21139217 CAUCCGCCGCCGCGAUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGACGGAGCUCACCGUCGAAUCGCUGGCCAAGAUGAGCAACUG--- ((((....((((((((..........(((((((.(((....))).))..(((....)))))))).(((((......)))))..))))).)))...))))........--- ( -39.80, z-score = -1.24, R) >droSec1.super_18 330652 107 + 1342155 CAUCCGCCGCCGCGGUACCAGUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGACGGAGCUCACCGUCGAAUCGCUGGCCAAGAUGAGCAACUG--- ..(((((((((..(((((........))))).)))))..((((.((((.....)))).)))).....))))((((((.((((......))))...).))))).....--- ( -43.60, z-score = -1.30, R) >droSim1.chr2L 20013963 107 + 22036055 CAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGACGGAGCUCACCGUCGAAUCGCUGGCCAAGAUGAGCAACUG--- ..(((((((((..(((((........))))).)))))..((((.((((.....)))).)))).....))))((((((.((((......))))...).))))).....--- ( -43.60, z-score = -1.73, R) >droPer1.super_10 1696102 109 + 3432795 -CUCCGCCGCCCCUGUACCAGUACCAGUACCAGGCGGAGACGCGGCUGACCACCAGUGGCGUGCAGACGGAGCUGACCGUGGAGUCGCUGGCCAAGCUAAGCAACAGCGC -((((.(((((...((((........))))..)))))..((((.((((.....)))).))))....((((......)))))))).(((((((........))..))))). ( -40.50, z-score = 0.38, R) >dp4.chr4_group4 2687700 109 + 6586962 -CUCCGCCGCCACUGUACCAGUACCAGUACCAGGCGGAGACGCGGCUGACCACCAGUGGCGUGCAGACGGAGCUGACCGUGGAGUCGCUGGCCAAGCUAAGCAACAGCGC -((((((((((((((.....(((....)))((((((....)))..))).....)))))))).))..((((......)))))))).(((((((........))..))))). ( -43.80, z-score = -0.56, R) >droAna3.scaffold_12916 3973828 98 + 16180835 ------CCGCCCGAAUACCG---UCAGUAUUCCGUGGAGACACGGCUGACCACCAGCGGUGUGCAGACGGAGCUCACGGCCGAGUCGCUGGCCAAGCUGAGCAACAG--- ------...........(((---((.((((.(((((....))).((((.....)))))).)))).))))).(((((.(((((......)))))....))))).....--- ( -41.00, z-score = -2.26, R) >droMoj3.scaffold_6500 9714858 90 - 32352404 -----AUCGCCGCCGCCC---------UAUCAGGCGGAGGUGCGCAUGACCAGCAGCGGUGUCCAGACGGAGCUGACAGUCGAGUCGCAGCUGAGUCAUAGCGG------ -----..((((.(((((.---------.....))))).))))((((((((((((.(((((.(((....))))))(((......))))).)))).))))).))).------ ( -41.90, z-score = -2.42, R) >consensus CAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGACGGAGCUCACCGUCGAAUCGCUGGCCAAGAUGAGCAACUG___ ..(((((((((..(((((........))))).)))))..((((.((((.....)))).)))).....))))(((((..((((......)))).....)))))........ (-29.80 = -30.83 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:22 2011