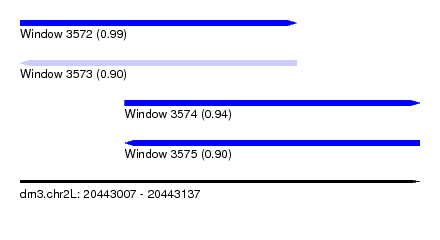
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,443,007 – 20,443,137 |
| Length | 130 |
| Max. P | 0.993049 |

| Location | 20,443,007 – 20,443,097 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.39114 |
| G+C content | 0.61002 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.96 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
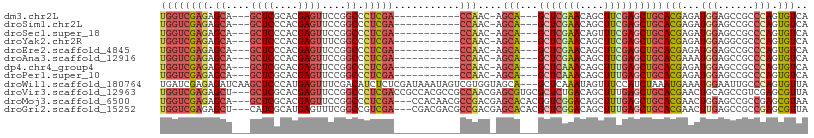
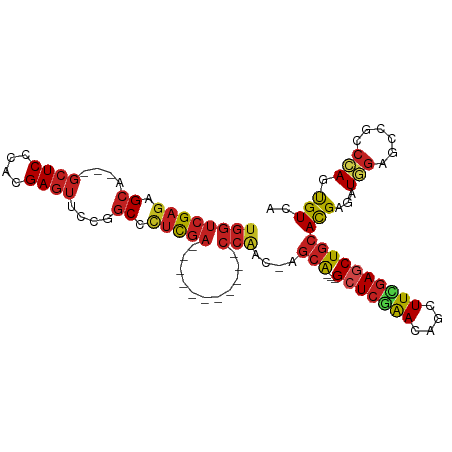
>dm3.chr2L 20443007 90 + 23011544 UGGUCGAGAGCA---GCUCGCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((.((((......))).).)). ( -42.10, z-score = -3.34, R) >droSim1.chr2L 20013719 90 + 22036055 UGGUCGAGAGCA---GCUCCCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((.((((......))).).)). ( -41.00, z-score = -3.61, R) >droSec1.super_18 330497 90 + 1342155 UGGUCGAGAGCA---GCUCCCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCGAACAGUUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((.((((......))).).)). ( -41.00, z-score = -4.05, R) >droYak2.chr2R 6808335 90 + 21139217 UGGUCGAGAGCA---GCUCCCACGAGUUUCGGCCCUCGA-----------CCAAC-AGCA---GCUCGAACAGCUUCGAGCUGCACGAGAUGGAGGCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..........(((((...))))). ( -41.50, z-score = -3.31, R) >droEre2.scaffold_4845 18599517 90 - 22589142 UGGUCGAGAGCA---GCUCCCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((.((((......))).).)). ( -41.00, z-score = -3.61, R) >droAna3.scaffold_12916 3973700 90 + 16180835 UGGUCGAGAGCA---GCUCCCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCGAACAGCUUCGAGCUGCACGAAAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((..(((......)))...)). ( -41.80, z-score = -4.30, R) >dp4.chr4_group4 2687580 90 + 6586962 UGGUCGAGAGCA---GCUCGCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCAAACAGCUUUGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((.((((......))).).)). ( -40.30, z-score = -3.36, R) >droPer1.super_10 1695982 90 + 3432795 UGGUCGAGAGCA---GCUCGCACGAGUUCCGGCCCUCGA-----------CCAAC-AGCA---GCUCAAACAGCUUUGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ((((((((.((.---((((....))))....)).)))))-----------)))..-.(((---(((((((....))))))))))..((.((((......))).).)). ( -40.30, z-score = -3.36, R) >droWil1.scaffold_180764 1855727 105 - 3949147 UGAUCGAGAGAUCAAGCUCCCAUGAGUUUCGACAUCUCUCGAUAAAUAGUCGUGGUAGCA---GCUCAAAUAGUUUCCAUCUAAAUGAAAUGGAAUUGCCCAGUGUUA (((((....))))).(((.((((((.(.((((......)))).....).)))))).)))(---((.........(((((((.....))..))))).........))). ( -27.07, z-score = -2.06, R) >droVir3.scaffold_12963 2607596 105 - 20206255 UGGUCGAGAGCU---GCUCGCACGAGUUCCGGCCCUCGACCGCCACGCCGCCAACGAGCGUGCGCUCUGACAGCUUUGAGCUGCACGAACUGCAGCCGUCGAGCGUUA .(((((((.(((---((((....))))...))).)))))))(((((((((....)).))))).))..((((.((((((.((((((.....))))))))..)))))))) ( -47.10, z-score = -1.91, R) >droMoj3.scaffold_6500 9714719 102 - 32352404 UGGUCGAGAGCA---GCUCGCACGAGUUCCGGCCCUCGA---CCACAACGCCGACGAGCACACGGUCGGACAGCUUUGAGCUGCACGAACUGGAGCCGCCGAGCGUAA ((((((((.((.---((((....))))....)).)))))---)))..((((((.((.((.(.((((((..((((.....))))..)).))))).)))).)).)))).. ( -46.10, z-score = -2.76, R) >droGri2.scaffold_15252 2171085 102 - 17193109 UGGUCGAGAGCU---CAUCGCAUGAGUUUCGGCCGUCGA---CGACGACGCCGACGAGCACACGCUCGGACAGCUUUGAGCUGCACGAACUUGAGCCGCCGAGCGUUA .((((((((.((---(((...)))))))))))))(((..---.)))(((((((.((.((.((.(.(((..((((.....))))..))).).)).)))).)).))))). ( -41.90, z-score = -1.47, R) >consensus UGGUCGAGAGCA___GCUCCCACGAGUUCCGGCCCUCGA___________CCAAC_AGCA___GCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCA ...(((((.((....((((....))))....)).)))))..................(((...(((((((....))))))))))(((...(((......))).))).. (-21.62 = -21.96 + 0.34)
| Location | 20,443,007 – 20,443,097 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.39114 |
| G+C content | 0.61002 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20443007 90 - 23011544 UGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGCGAGC---UGCUCUCGACCA .((((.((((....))))......((((((((((....)))))))---))))-)))((-----------((((((((.((..(((....))))---))))))))))). ( -41.50, z-score = -1.70, R) >droSim1.chr2L 20013719 90 - 22036055 UGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGGGAGC---UGCUCUCGACCA .......(((((((((((((....((((((((((....)))))))---))).-...))-----------)((((........))))..)))))---)))))....... ( -42.10, z-score = -1.87, R) >droSec1.super_18 330497 90 - 1342155 UGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAACUGUUCGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGGGAGC---UGCUCUCGACCA .......(((((((((((((....((((((((((....)))))))---))).-...))-----------)((((........))))..)))))---)))))....... ( -42.20, z-score = -2.55, R) >droYak2.chr2R 6808335 90 - 21139217 UGACACUGGGCGCCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGC---UGCU-GUUGG-----------UCGAGGGCCGAAACUCGUGGGAGC---UGCUCUCGACCA .((((.((((....))))......((((((((((....)))))))---))))-)))((-----------((((((((.(...(((....))))---.)))))))))). ( -40.60, z-score = -1.98, R) >droEre2.scaffold_4845 18599517 90 + 22589142 UGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGGGAGC---UGCUCUCGACCA .......(((((((((((((....((((((((((....)))))))---))).-...))-----------)((((........))))..)))))---)))))....... ( -42.10, z-score = -1.87, R) >droAna3.scaffold_12916 3973700 90 - 16180835 UGACACUGGGCGGCUCCAUUUCGUGCAGCUCGAAGCUGUUCGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGGGAGC---UGCUCUCGACCA .......((((((((((....((.((((((((((....)))))))---))).-.((((-----------(.....)))))....))..)))))---)))))....... ( -41.50, z-score = -1.95, R) >dp4.chr4_group4 2687580 90 - 6586962 UGACACUGGGCGGCUCCAUCUCGUGCAGCUCAAAGCUGUUUGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGCGAGC---UGCUCUCGACCA .((((.((((....))))......((((((((((....)))))))---))))-)))((-----------((((((((.((..(((....))))---))))))))))). ( -39.70, z-score = -1.76, R) >droPer1.super_10 1695982 90 - 3432795 UGACACUGGGCGGCUCCAUCUCGUGCAGCUCAAAGCUGUUUGAGC---UGCU-GUUGG-----------UCGAGGGCCGGAACUCGUGCGAGC---UGCUCUCGACCA .((((.((((....))))......((((((((((....)))))))---))))-)))((-----------((((((((.((..(((....))))---))))))))))). ( -39.70, z-score = -1.76, R) >droWil1.scaffold_180764 1855727 105 + 3949147 UAACACUGGGCAAUUCCAUUUCAUUUAGAUGGAAACUAUUUGAGC---UGCUACCACGACUAUUUAUCGAGAGAUGUCGAAACUCAUGGGAGCUUGAUCUCUCGAUCA ........((((.((((((((.....)))))))).(.....)...---)))).............(((((((((..((((..(((....))).))))))))))))).. ( -24.30, z-score = -1.07, R) >droVir3.scaffold_12963 2607596 105 + 20206255 UAACGCUCGACGGCUGCAGUUCGUGCAGCUCAAAGCUGUCAGAGCGCACGCUCGUUGGCGGCGUGGCGGUCGAGGGCCGGAACUCGUGCGAGC---AGCUCUCGACCA ...((((((((((((..((((.....))))...))))))).)))))(((((.((....)))))))..((((((((((.(...(((....))))---.)))))))))). ( -51.70, z-score = -1.66, R) >droMoj3.scaffold_6500 9714719 102 + 32352404 UUACGCUCGGCGGCUCCAGUUCGUGCAGCUCAAAGCUGUCCGACCGUGUGCUCGUCGGCGUUGUGG---UCGAGGGCCGGAACUCGUGCGAGC---UGCUCUCGACCA ..((((.(((((((....(.(((..((((.....))))..))).)....)).)))))))))..(((---((((((((.((..(((....))))---)))))))))))) ( -49.20, z-score = -2.39, R) >droGri2.scaffold_15252 2171085 102 + 17193109 UAACGCUCGGCGGCUCAAGUUCGUGCAGCUCAAAGCUGUCCGAGCGUGUGCUCGUCGGCGUCGUCG---UCGACGGCCGAAACUCAUGCGAUG---AGCUCUCGACCA ....(.((((.((((((.(((((..((((.....))))..)))))(((((....(((((((((...---.)))).)))))....)))))..))---)))).)))).). ( -47.80, z-score = -2.96, R) >consensus UGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGC___UGCU_GUUGG___________UCGAGGGCCGGAACUCGUGCGAGC___UGCUCUCGACCA ........((((((.....((((.(((((.....))))).)))).....))).))).............((((((((.....(((....))).....))))))))... (-25.34 = -25.67 + 0.33)

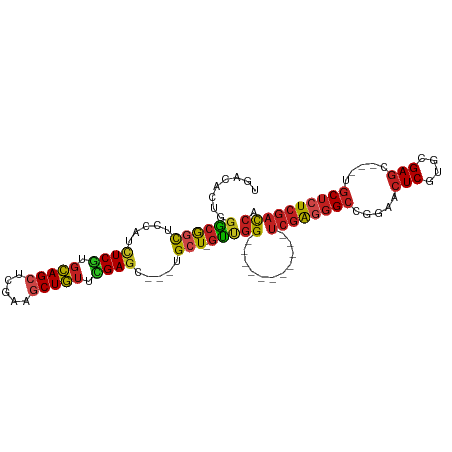

| Location | 20,443,041 – 20,443,137 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.57553 |
| G+C content | 0.63377 |
| Mean single sequence MFE | -38.61 |
| Consensus MFE | -16.86 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20443041 96 + 23011544 ---------------GACCAACAGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGUGCCGCCUCCACUGGUUUCCAGUCA ---------------(((.....((((((((((....))))))))))((.((.(((((((((((((......)))...)))))(((...)))))))))).)).....))). ( -41.90, z-score = -2.47, R) >droSim1.chr2L 20013753 96 + 22036055 ---------------GACCAACAGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGGGCCGCCUCCACUGGUUGCCAGUCA ---------------(((((((.((((((((((....))))))))))..(((.(((..((((((...(((........))))))))).))).))).....))))...))). ( -43.90, z-score = -2.12, R) >droSec1.super_18 330531 96 + 1342155 ---------------GACCAACAGCAGCUCGAACAGUUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGUGCCGCCUCCACUGGUUGCCAGUCA ---------------(((((((.((((((((((....))))))))))...((.(((((((((((((......)))...)))))(((...)))))))))).))))...))). ( -42.10, z-score = -2.32, R) >droYak2.chr2R 6808369 96 + 21139217 ---------------GACCAACAGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGGCGCCCAGUGUCAUCUGCAAGGCGGGGGUGCCGCCUCCACUGGUUGCCAGUCA ---------------(((((((.((((((((((....))))))))))...((.(((((((((((..((((........))))..)).).)))))))))).))))...))). ( -46.60, z-score = -2.73, R) >droEre2.scaffold_4845 18599551 96 - 22589142 ---------------GACCAACAGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGGGCGCCGCCUUCCCUGGUUGUCAGUCA ---------------(((..(((((((((((((....))))))))))((.((..(((.((((((((......)))...)))))((((...))))))))).)))))..))). ( -40.80, z-score = -1.40, R) >droAna3.scaffold_12916 3973734 82 + 16180835 ---------------GACCAACAGCAGCUCGAACAGCUUCGAGCUGCACGAAAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGCCGCCGCCACCACCA-------------- ---------------((((....((((((((((....))))))))))......(((......)))).)))........((((....)))).......-------------- ( -29.00, z-score = -2.12, R) >dp4.chr4_group4 2687614 89 + 6586962 ---------------GACCAACAGCAGCUCAAACAGCUUUGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGUGCCCCCCUGGCCGCGACUC------- ---------------........((((((((((....))))))))))........(((.(((((((.(.(((((((...))))))).)...))))..))).)))------- ( -33.60, z-score = -0.95, R) >droPer1.super_10 1696016 89 + 3432795 ---------------GACCAACAGCAGCUCAAACAGCUUUGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGUGCCCCCCUGGCCGCGACUC------- ---------------........((((((((((....))))))))))........(((.(((((((.(.(((((((...))))))).)...))))..))).)))------- ( -33.60, z-score = -0.95, R) >droVir3.scaffold_12963 2607630 105 - 20206255 GACCGCCACGCCGCCAACGAGCGUGCGCUCUGACAGCUUUGAGCUGCACGAACUGCAGCCGUCGAGCGUUAUAUGCAAGGCGGCGGCGCCACCAGCCAGGCUGGA------ ..(((((..((((((...((((....))))((((.((((((.((((((.....))))))))..)))))))).......))))))(((.......))).))).)).------ ( -48.30, z-score = -1.71, R) >droMoj3.scaffold_6500 9714753 99 - 32352404 ---GACCACAACGCCGACGAGCACACGGUCGGACAGCUUUGAGCUGCACGAACUGGAGCCGCCGAGCGUAAUAUGCAAGGCGGCACCCAUGCUGGACUAUCA--------- ---..............(.((((..((((((..((((.....))))..)).))))..((((((..((((...))))..)))))).....)))).).......--------- ( -36.30, z-score = -2.28, R) >droGri2.scaffold_15252 2171119 94 - 17193109 ---GACGACGACGCCGACGAGCACACGCUCGGACAGCUUUGAGCUGCACGAACUUGAGCCGCCGAGCGUUAUCUGUAAAGCGGCACCACCACCA-CCA------------- ---...((..((((((.((.((.((.(.(((..((((.....))))..))).).)).)))).)).))))..))........((.....))....-...------------- ( -28.60, z-score = -1.54, R) >consensus _______________GACCAACAGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGGGCGCCGCCUCCCCUGGUUG_______ ..........................(((((((....)))))))..............(((((..((.......))..)))))............................ (-16.86 = -16.43 + -0.44)


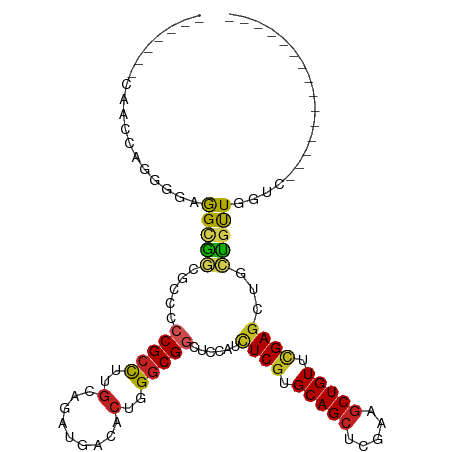
| Location | 20,443,041 – 20,443,137 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.57553 |
| G+C content | 0.63377 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20443041 96 - 23011544 UGACUGGAAACCAGUGGAGGCGGCACGCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCUGUUGGUC--------------- .(((..(...(((((((((((((.....))))))).(....).))))))((((.......))))((((((((((....))))))))))..)..)))--------------- ( -41.70, z-score = -1.44, R) >droSim1.chr2L 20013753 96 - 22036055 UGACUGGCAACCAGUGGAGGCGGCCCGCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCUGUUGGUC--------------- .(((..(((.(((((((((((((.....))))))).(....).))))))((((.......))))((((((((((....)))))))))))))..)))--------------- ( -46.20, z-score = -2.15, R) >droSec1.super_18 330531 96 - 1342155 UGACUGGCAACCAGUGGAGGCGGCACGCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAACUGUUCGAGCUGCUGUUGGUC--------------- .(((..(((.(((((((((((((.....))))))).(....).))))))((((.......))))((((((((((....)))))))))))))..)))--------------- ( -46.30, z-score = -2.89, R) >droYak2.chr2R 6808369 96 - 21139217 UGACUGGCAACCAGUGGAGGCGGCACCCCCGCCUUGCAGAUGACACUGGGCGCCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCUGUUGGUC--------------- .(((..(((((.(((((((((((.....))(((...(((......))))))))))))).)).))((((((((((....)))))))))))))..)))--------------- ( -47.40, z-score = -3.08, R) >droEre2.scaffold_4845 18599551 96 + 22589142 UGACUGACAACCAGGGAAGGCGGCGCCCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCUGUUGGUC--------------- .(((..(((....((((.((.(.(((((..((...))((......))))))).).)).))))..((((((((((....)))))))))))))..)))--------------- ( -44.00, z-score = -2.14, R) >droAna3.scaffold_12916 3973734 82 - 16180835 --------------UGGUGGUGGCGGCGGCGCCUUGCAGAUGACACUGGGCGGCUCCAUUUCGUGCAGCUCGAAGCUGUUCGAGCUGCUGUUGGUC--------------- --------------....((..(((((((.(((.(.(((......))).).))).))...(((.(((((.....))))).))))))))..))....--------------- ( -33.80, z-score = -0.47, R) >dp4.chr4_group4 2687614 89 - 6586962 -------GAGUCGCGGCCAGGGGGGCACCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCAAAGCUGUUUGAGCUGCUGUUGGUC--------------- -------(((((((...(((((((....)).)))))(((......))).))))))).(((....((((((((((....))))))))))....))).--------------- ( -38.40, z-score = -0.69, R) >droPer1.super_10 1696016 89 - 3432795 -------GAGUCGCGGCCAGGGGGGCACCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCAAAGCUGUUUGAGCUGCUGUUGGUC--------------- -------(((((((...(((((((....)).)))))(((......))).))))))).(((....((((((((((....))))))))))....))).--------------- ( -38.40, z-score = -0.69, R) >droVir3.scaffold_12963 2607630 105 + 20206255 ------UCCAGCCUGGCUGGUGGCGCCGCCGCCUUGCAUAUAACGCUCGACGGCUGCAGUUCGUGCAGCUCAAAGCUGUCAGAGCGCACGCUCGUUGGCGGCGUGGCGGUC ------.(((((...)))))....((((((((...((......(((.((((((((((.((((..(((((.....)))))..))))))).)).)))))))))))))))))). ( -48.30, z-score = -0.46, R) >droMoj3.scaffold_6500 9714753 99 + 32352404 ---------UGAUAGUCCAGCAUGGGUGCCGCCUUGCAUAUUACGCUCGGCGGCUCCAGUUCGUGCAGCUCAAAGCUGUCCGACCGUGUGCUCGUCGGCGUUGUGGUC--- ---------.....(.((((((((((.((((((..((.......))..)))))).))...(((..((((.....))))..))).)))))(((....)))....))).)--- ( -39.00, z-score = -1.36, R) >droGri2.scaffold_15252 2171119 94 + 17193109 -------------UGG-UGGUGGUGGUGCCGCUUUACAGAUAACGCUCGGCGGCUCAAGUUCGUGCAGCUCAAAGCUGUCCGAGCGUGUGCUCGUCGGCGUCGUCGUC--- -------------((.-.(((((.....)))))...))((..((((.(((((((.((.(((((..((((.....))))..))))).)).)).)))))))))..))...--- ( -45.50, z-score = -3.83, R) >consensus _______CAACCAGGGGAGGCGGCGCCCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCUGUUGGUC_______________ ..................(((((.....(((((..(.........)..)))))......((((.(((((.....))))).))))...)))))................... (-18.81 = -18.58 + -0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:21 2011