| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,421,779 – 20,421,845 |
| Length | 66 |
| Max. P | 0.997251 |

| Location | 20,421,779 – 20,421,845 |
|---|---|
| Length | 66 |
| Sequences | 10 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 66.99 |
| Shannon entropy | 0.69655 |
| G+C content | 0.35714 |
| Mean single sequence MFE | -17.03 |
| Consensus MFE | -4.56 |
| Energy contribution | -4.12 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
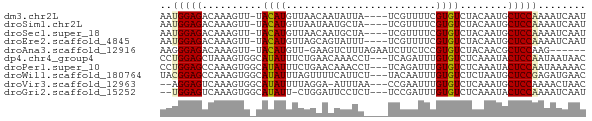
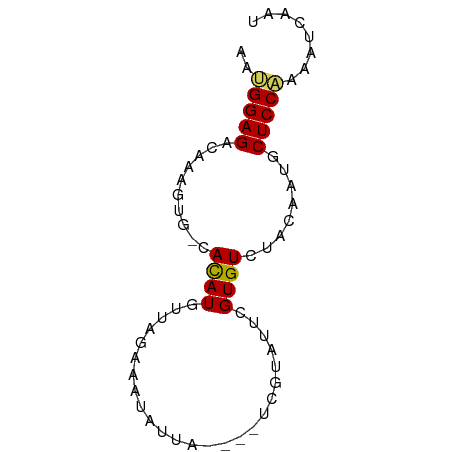
>dm3.chr2L 20421779 66 + 23011544 AAUGGAGACAAAGUU-UACAUGUUAACAAUAUUA----UCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAU ..(((((.((..((.-.(((((..(((.......----..)))..)))))..))..)))))))........ ( -15.80, z-score = -3.78, R) >droSim1.chr2L 19991860 66 + 22036055 AAUGGAGACAAAGUU-UACAUGUUAAUAAUGCUA----UCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAU ..(((((.((..((.-.(((((..(((.......----..)))..)))))..))..)))))))........ ( -13.80, z-score = -2.63, R) >droSec1.super_18 309482 66 + 1342155 AAUGGAGACAAAGUU-UACAUGUUAACAAUGCUA----UCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAU ..(((((.((..((.-.(((((..(((.......----..)))..)))))..))..)))))))........ ( -15.80, z-score = -3.29, R) >droEre2.scaffold_4845 18578841 66 - 22589142 AAUGGAGACAAAGUU-UACAUGUUAGCAGUAUUU----UCGUUUUCGUGUCUACAAUGCUCCAAAAUCAAU ..(((((.((..((.-.(((((..(((.......----..)))..)))))..))..)))))))........ ( -15.80, z-score = -3.25, R) >droAna3.scaffold_12916 3954347 63 + 16180835 AAGGGAGACAAAGUU-UACAUGUU-GAAGUCUUUAGAAUCUUCUCCGUGUCUACAACGCUCCAAG------ ...((((.....((.-.(((((..-((((((....))..))))..)))))..))....))))...------ ( -15.90, z-score = -1.89, R) >dp4.chr4_group4 2668894 68 + 6586962 CCUGGAGCUAAAGUGGCAUAUUUCUGAACAAACCU---UCAGAUUUGUGUCUCAAAUACUCCAAUAAUAAC ..(((((.......((((((..((((((......)---)))))..)))))).......)))))........ ( -20.74, z-score = -4.88, R) >droPer1.super_10 1677258 68 + 3432795 CCUGGAGCCAAAGUGGCAUAUUUCUGAACAAACCU---UCAGAUUUGUGUCUCAAAUACUCCAAUAAAAAC ..(((((.......((((((..((((((......)---)))))..)))))).......)))))........ ( -20.74, z-score = -4.81, R) >droWil1.scaffold_180764 2080462 68 + 3949147 UACGGAGCCAAAGUGGCAUAUUUAGUUUUCAUUCU---UACAAUUUGUGUCUCUAAUGCUCCGAGAUGAAC ..((((((...((.((((((..(.((.........---.)).)..)))))).))...))))))........ ( -18.20, z-score = -2.77, R) >droVir3.scaffold_12963 2584656 65 - 20206255 --AGGAGUCAAAGUGGCAUAUUUUAGGA-AUUUAA---CCGAAUUUGUGUCUCAAAUGCUCCAAAACUAAC --.(((((....(.((((((..((.((.-......---)).))..)))))).)....)))))......... ( -15.70, z-score = -2.58, R) >droGri2.scaffold_15252 2147628 65 - 17193109 --UGGAGUCAAAGUGGCAUAUU-CUGGAUUCCUCU---UCCGAUUUGUGUCUCAAAUACUCCAAAAUCAAU --((((((....(.((((((..-.((((.......---))))...)))))).)....))))))........ ( -17.80, z-score = -3.32, R) >consensus AAUGGAGACAAAGUG_CACAUGUUAGAAAUAUUA____UCGUAUUCGUGUCUACAAUGCUCCAAAAUCAAU ..(((((.........(((...........................))).........)))))........ ( -4.56 = -4.12 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:17 2011