| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,421,605 – 20,421,688 |
| Length | 83 |
| Max. P | 0.999883 |

| Location | 20,421,605 – 20,421,688 |
|---|---|
| Length | 83 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 61.38 |
| Shannon entropy | 0.86546 |
| G+C content | 0.34481 |
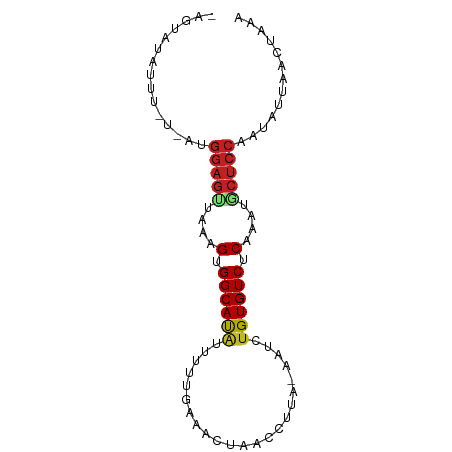
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -9.55 |
| Energy contribution | -9.81 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.70 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
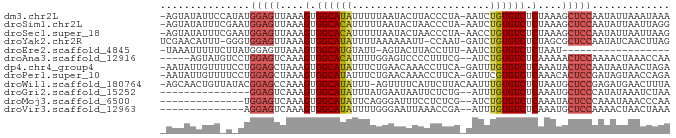
>dm3.chr2L 20421605 83 + 23011544 -AGUAUAUUCCAUAUGGAGUUAAAGUGGCAUAUUUUUAAUACUUACCCUA-AAUCUGUGUCUCUAAAGCUCCAAUAUUAAAUAAA -....((((..((((((((((..((.((((((..((((..........))-))..)))))).))..)))))).))))..)))).. ( -18.10, z-score = -3.47, R) >droSim1.chr2L 19991686 83 + 22036055 -AGUAUAUUUCGAAUGGAGUUAAAGUGGCACAUUUUUAAUACUAACCCUA-AAUCUGUGUCUCUAAAGCUCCAAUAUUAAUUAGG -.............(((((((..((.((((((..((((..........))-))..)))))).))..)))))))............ ( -19.30, z-score = -3.01, R) >droSec1.super_18 309309 83 + 1342155 -AGUAUAUUUCGAAUGGAGUUAAAGUGGCACAUUUUUAAUACUAACCCUA-AACCUGUGUCUCUAAAGCUCCAAUAUUAAUUAAG -.............(((((((..((.((((((..((((..........))-))..)))))).))..)))))))............ ( -18.50, z-score = -3.23, R) >droYak2.chr2R 6785337 82 + 21139217 UCGAACAUUU-GGGUGGAGUUAAAGUGGCAUAUUUUAAAAAAUU-CCAAU-GAUCUGUGUCUCUAGCGCUCCAAUAUCAACUUAG ........((-((.((((((...((.((((((..(((.......-....)-))..)))))).))...))))))...))))..... ( -17.30, z-score = -1.59, R) >droEre2.scaffold_4845 18578976 64 - 22589142 -UAAAUUUUUCUUAUGGAGUUAAAGUGGCAUGUAUU-AGUACUUACCUUU-AAUCUGUGUCUCUAAU------------------ -..((((((......))))))..((.(((((..(((-((.........))-)))..))))).))...------------------ ( -7.70, z-score = 0.01, R) >droAna3.scaffold_12916 3954809 78 + 16180835 -----AGUAUGUCCUGGAGUCAAAGUGGCACAUUUUGGAGUCCCCUUUCG--AUCUGUGUCUCAAAAACUCCAAAACUAAACCAA -----.....((..((((((....(.((((((..((((((.....)))))--)..)))))).)....))))))..))........ ( -20.40, z-score = -2.67, R) >dp4.chr4_group4 2668718 83 + 6586962 -AAUAUUGUUUUCCUGGAGCUAAAGUGGCAUAUUUCUGAACAAACCUUCA-GAUUUGUGUCUCAAAUACUCCAAUAAUAACUAGA -..((((((.....(((((.......((((((..((((((......))))-))..)))))).......)))))))))))...... ( -21.64, z-score = -3.77, R) >droPer1.super_10 1677082 83 + 3432795 -AAUAUUGUUUUCCUGGAGCUAAAGUGGCAUAUUUCUGAACAAACCUUCA-GAUUCGUGUCUCAAACACUCCGAUAGUAACCAGA -..(((((((.....((((.......(((((...((((((......))))-))...))))).......)))))))))))...... ( -18.24, z-score = -1.77, R) >droWil1.scaffold_180764 2080278 83 + 3949147 -AGCAACUGUUAUACGGAGCCAAAGUGGCAUAUUU-AGUUUUCAUUCUUACAAUUUGUGUCUCUAAUGCUCCGAGAUGAACUUUA -.............((((((...((.((((((..(-.((..........)).)..)))))).))...))))))............ ( -18.20, z-score = -1.44, R) >droGri2.scaffold_15252 2147476 68 - 17193109 ---------------GGAGUCAAAGUGGCAUAUUUAUGAAUAAUUCUCUG--AUUUGUGUCUCAAAUGCUCCCAUAUAAAUCUAA ---------------(((((....(.((((((.....((.......))..--...)))))).)....)))))............. ( -12.10, z-score = -1.37, R) >droMoj3.scaffold_6500 9689782 69 - 32352404 --------------UGGAGUCAAAGUGGCAUAUUCAGGGAUUUCCUCUCG--AUCUGUGUCUCAAAUACUCCCAAAUAAACCCAA --------------.(((((....(.((((((.((((((.....)))).)--)..)))))).)....)))))............. ( -15.70, z-score = -1.51, R) >droVir3.scaffold_12963 2584503 69 - 20206255 --------------AGGAGUCAAAGUGGCAUAUUUUGGGAAUUAAACCGA--AUUUGUGUCUCAAAUGCUCCAAAACUAACUAAA --------------.(((((....(.((((((.(((((........))))--)..)))))).)....)))))............. ( -19.00, z-score = -3.35, R) >consensus _AGUAUAUUU_U_AUGGAGUUAAAGUGGCAUAUUUUUGAAACUAACCUUA_AAUCUGUGUCUCAAAUGCUCCAAUAUUAACUAAA ...............(((((....(.((((((.......................)))))).)....)))))............. ( -9.55 = -9.81 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:16 2011