| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,419,457 – 20,419,613 |
| Length | 156 |
| Max. P | 0.973744 |

| Location | 20,419,457 – 20,419,613 |
|---|---|
| Length | 156 |
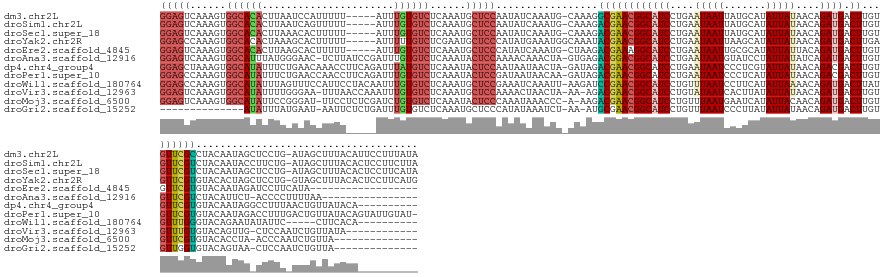
| Sequences | 12 |
| Columns | 163 |
| Reading direction | forward |
| Mean pairwise identity | 69.28 |
| Shannon entropy | 0.66665 |
| G+C content | 0.37090 |
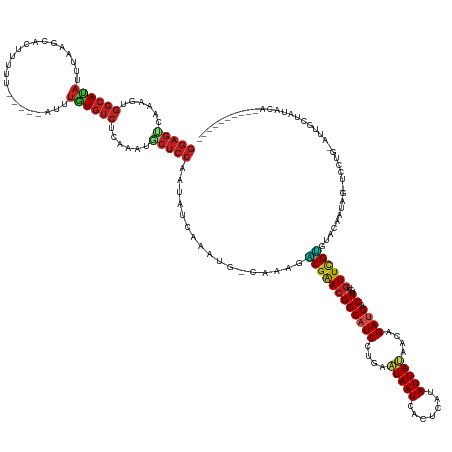
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20419457 156 + 23011544 GGAGUCAAAGUGGCACACUUAAUCCAUUUUU-----AUUUGUGUCUCAAAUGCUCCAAUAUCAAAUG-CAAAGGCGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGCCUACAAUAGCUCCUG-AUAGCUUUACAUUCCUUUAUA (((((....(.((((((..(((.......))-----)..)))))).)....)))))..(((((...(-(..((((((((((((((.((.(((((.......)))))..)))))).))...))))))))......))...))-))).................. ( -36.40, z-score = -1.96, R) >droSim1.chr2L 19989487 156 + 22036055 GGAGUCAAAGUGGCACACUUAAUCAGUUUUU-----AUUUGUGUCUCAAAUGCUCCAAUAUCAAAUG-CAAAGACGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGUCUACAAUACCUUCUG-AUAGCUUUACACUCCUUCUUA (((((.(((((((((((..(((.......))-----)..))))))(((..(((.............)-)).((((((((((((((.((.(((((.......)))))..)))))).))...))))))))...........))-)..)))))..)))))...... ( -34.42, z-score = -1.96, R) >droSec1.super_18 307086 156 + 1342155 GGAGUCAAAGUGGCACACUUAAACACUUUUU-----AUUUGUGUCUCAAAUGCUCCAAUAUCAAAUG-CAAAGACGAACGGCAUCCUGAAUAAUUAUGCAUAUUAUAACAGAUGACUUGUGUUCGUCUACAAUAGCUCCUG-AUAGCUUUACACUCCUUCAUA (((((.(((((((((((..((((.....)))-----)..)))))).............(((((...(-(..((((((((((((((.((.(((((.......)))))..)))))).))...))))))))......))...))-))))))))..)))))...... ( -36.30, z-score = -2.51, R) >droYak2.chr2R 6785651 157 + 21139217 GGAGCCAAAGUGGCACACUAAAGCACUUUUU-----AUUUUUGUCUCGAAUGCUCCCAUAUGAAAUGGCAAAUACGAACGGCAUCCUGAAUAAUUAAGCAUAUUAUAACAGAUGACUUGAGUUCGUGUACACUAGCUCCUG-GUAGCUUUACACUCCUUCAUG ((((((.....)))....((((((.......-----....((((((((.(((....))).)))...)))))((((((((((((((.((.(((((.......)))))..)))))).))...))))))))..(((((...)))-)).))))))...)))...... ( -32.60, z-score = -0.23, R) >droEre2.scaffold_4845 18576995 139 - 22589142 GGAGUCAAAGUGGCACACUUAAGCACUUUUU-----AUUUGUGUCUCAAAUGCUCCCAUAUCAAAUG-CUAAGACGAAAGGCAUCCUGAAUAAUUGCGCAUAUUAUUACAGAUGACUUGUGUUCGUGUACAAUAGAUCCUUCAUA------------------ (((((....(.((((((..((((.....)))-----)..)))))).)....)))))...........-.......(((.((.(((.((.(((...(((((..(((((...)))))..)))))...))).))...))))))))...------------------ ( -27.80, z-score = 0.12, R) >droAna3.scaffold_12916 3954510 144 + 16180835 GGAGUCAAAGUGGCAUUUAUGGGAAC-UCUUAUCCGAUUUGUGUCUCAAAUACUCCAAAACAAACUA-GUGAGACGGACGGCAUCCUGAAUAAUGUAUCCUAUUAUAUCAGAUGACUUGUGUUCGUCUACAUUCU-ACCCCUUUUAA---------------- (((((....(.(((((..((.(((..-.....))).))..))))).)....)))))..........(-(((((((((((((((((.(((((((((.....)))))).))))))).))...)))))))).))))..-...........---------------- ( -37.80, z-score = -2.38, R) >dp4.chr4_group4 2666651 152 + 6586962 GGAGCUAAAGUGGCAUAUUUCUGAACAAACCUUCAGAUUUAUGUCUCAAAUACUCCAAUAAUAACUA-GAUAGACGAACGGCAUCCUGAAUAAUCCCUCGUAUUAUAACAGACGACUUGUGUUCGUGUACAAUAGGCCUUUAACUGUUAUACA---------- ((((.......((((((..((((((......))))))..)))))).......))))...........-.....((((((((.((........)).))((((..........)))).....))))))(((.(((((........))))).))).---------- ( -34.04, z-score = -1.96, R) >droPer1.super_10 1674849 161 + 3432795 GGAGCCAAAGUGGCAUAUUUCUGAACCAACCUUCAGAUUUGUGUCUCAAAUACUCCGAUAAUAACAA-GAUAGACGAACGGCAUCCUGAAUAAUCCCUCAUAUUAUAACAGACGACUUGUGUUCGUGUACAAUAGACCUUUGACUGUUAUACAGUAUUGUAU- ((((.......((((((..((((((......))))))..)))))).......)))).((((((....-.....(((((((.(((((((.(((((.......)))))..)))..))..)))))))))(((.(((((........))))).)))..))))))..- ( -34.74, z-score = -1.05, R) >droWil1.scaffold_180764 2077498 147 + 3949147 GGAGCCAAAGUGGCAUAUUUAGUUUCCAUUCCUACAAUUUGUGUCUCAAAUGCUCCGAAAUCAAAUU-AAGAUCCGAACGGCAUCCUGUUUAAUCCUUCAUAUUAAAACAGAUGACUUAUGUUUGGGUACAGAAUAUAUUC-----CUUCACA---------- (((((....(.((((((..(.((..........)).)..)))))).)....)))))(((........-...(((((((((.((((.(((((..............))))))))).....)))))))))...(((....)))-----.)))...---------- ( -29.44, z-score = -0.94, R) >droVir3.scaffold_12963 2581638 147 - 20206255 GGAGUCAAAGUGGCAUAUUUUGGGAA-UUUAACCAAAUUUGUGUCUCAAAUGCUCCAAAACUAACUA-AA-AGACGAACGGCAUCCUGUAUAAUCACUUAUAUUAUAACAGAUGACUUGUGUUUGUGUACAGUUG-CUCCAAUCUGUUAUA------------ (((((....(.((((((.(((((...-.....)))))..)))))).)....))))).........((-(.-(((.....((((..(((((((..(((...((((......))))....)))....))))))).))-))....))).)))..------------ ( -33.40, z-score = -1.49, R) >droMoj3.scaffold_6500 9688259 145 - 32352404 GGAGUCAAAGUGGCAUAUUCCGGGAU-UUCCUCUCGAUCUGUGUCUCAAAUACUCCCAAAUAAACCC-A-AAGACGAACGGCAUCCUGUUUAAUGAAUCAUAUUACAACAGAUGACUUGUGUUCGUGUACACCUA-ACCCAAUCUGUUA-------------- (((((....(.((((((...(((((.-....)))))...)))))).)....)))))...........-.-.(.((((((((((((.(((((((((.....))))).)))))))).))...)))))).).......-.............-------------- ( -30.70, z-score = -0.49, R) >droGri2.scaffold_15252 2147490 131 - 17193109 --------------AUAUUUAUGAAU-AAUUCUCUGAUUUGUGUCUCAAAUGCUCCCAUAUAAAUCU-AA-AUGCGAACGGCAUCCUGUUUAAUCCCUUAUAUUAUAACAGAUGACUUGUGUUGGUGUACAGUAA-CUCCAAUCUGUUA-------------- --------------.......(((((-(.......(((((((((.............))))))))).-..-((((.....))))..)))))).............(((((((((((....)))((.((......)-).)).))))))))-------------- ( -19.82, z-score = -0.21, R) >consensus GGAGUCAAAGUGGCAUAUUUAAGCACUUUUU_____AUUUGUGUCUCAAAUGCUCCAAUAUCAAAUG_CAAAGACGAACGGCAUCCUGAAUAAUCACUCAUAUUAUAACAGAUGACUUGUGUUCGUGUACAAUAG_UCCUG_AUUGCUAUACA__________ (((((......((((((......................))))))......))))).................((((((((((((....(((((.......)))))....)))).))...))))))..................................... (-17.58 = -17.95 + 0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:15 2011