| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,419,148 – 20,419,284 |
| Length | 136 |
| Max. P | 0.986803 |

| Location | 20,419,148 – 20,419,284 |
|---|---|
| Length | 136 |
| Sequences | 12 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 73.80 |
| Shannon entropy | 0.57802 |
| G+C content | 0.37990 |
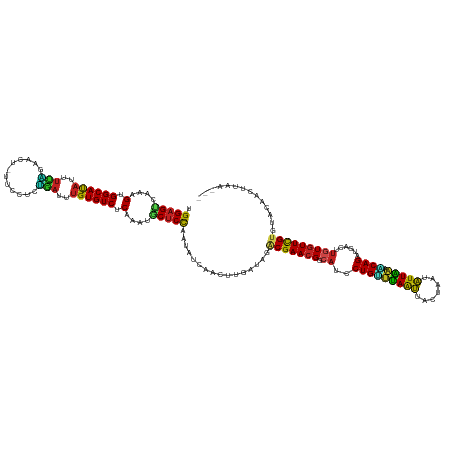
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -22.34 |
| Energy contribution | -21.09 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
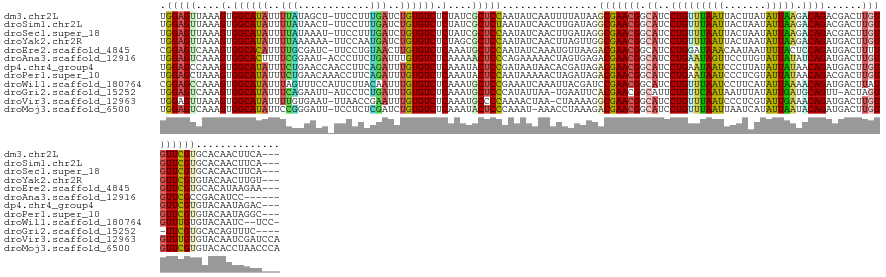
>dm3.chr2L 20419148 136 + 23011544 UGGAGUUAAAGUGGCAUAUUUUAUAGCU-UUCCUUUGAUCUGUGUCUCUAUCGCUCCAAUAUCAAUUUUAUAAGCGAACGGCAUCCUGUUUAAUUACUUAUAUUAAGACAGACGACUUGUGUUCGUGCACAACUUCA--- ((((((.(.((.((((((..(((.((..-...)).)))..)))))).)).).))))))...............(((((((.(((((((((((((.......))).))))))..))..)))))))))...........--- ( -28.40, z-score = -1.05, R) >droSim1.chr2L 19989183 136 + 22036055 UGGAGUUAAAGUGGCAUAUUUUAUAACU-UUCCUUUGAUCUGUGUCUCUAUCGCUCUAAUAUCAACUUGAUAGGCGAACGGCAUCCUGUUUAAUUACUAAUAUUAAGACAGACGACUUGUGUUCGUGCACAACUUCA--- .((((((..((.((((((..(((.....-......)))..)))))).)).(((((....((((.....)))))))))...((...(((((((((.......))).))))))(((((....).))))))..)))))).--- ( -28.00, z-score = -0.62, R) >droSec1.super_18 306782 136 + 1342155 UGGAGUUAAAGUGGCAUAUUUUAUAAAU-UUCCUUUGAUCUGUGUCUCUAUCGCUCCAAUAUCAACUUGAUAGGCGAACGGCAUCCUGUUUAAUUACUAAUAUUAAGACAGACGACUUGUGUUCGUGCACAACUUCA--- ((((((.(.((.((((((..(((.....-......)))..)))))).)).).)))))).((((.....)))).(((((((.(((((((((((((.......))).))))))..))..)))))))))...........--- ( -28.90, z-score = -0.84, R) >droYak2.chr2R 6785350 136 + 21139217 UGGAGUUAAAGUGGCAUAUUUUAAAAAA-UUCCAAUGAUCUGUGUCUCUAGCGCUCCAAUAUCAACUUAGUUGGCGAACGGCAUCCUGUUUAAUUACUAAUAUUAAGACAGAUGACUUGUGUUCGUGUACAACUUGU--- ((((((...((.((((((..(((.....-......)))..)))))).))...))))))......((..(((((((((((((((((.((((((((.......))).))))))))).))...))))))...))))).))--- ( -31.30, z-score = -1.33, R) >droEre2.scaffold_4845 18576060 136 - 22589142 CGGAGUCAAAGUGGCACAUUUUGCGAUC-UUCCUGUAACUUGUGUCUCAAAUGCUCCAAUAUCAAAUGUUAAGACGAACGGCAUCCUGGAUAAACAAUAAUUUUACUCCAGAUGACUUUUGUUCGUGCACAUAAGAA--- .(((((....(.((((((..(((((...-....)))))..)))))).)....)))))....((..((((....((((((((((((.((((((((.......)))).)))))))).))...))))))..))))..)).--- ( -35.50, z-score = -2.39, R) >droAna3.scaffold_12916 3954193 133 + 16180835 UGGAGUCAAAGUGGCACUUUUCGGAAU-ACCCUUCUGAUUUGUGUCUCAAAAACUCCCAGAAAAACUAGUGAGACGAACGGCAUCCUGAAUAGUUCCUUGUAUUAUAUCAGAUGACUUGUGUUCGCCGACAUCC------ .((((((...(.(((((...((((((.-....))))))...))))).).....((((.((.....)).).))).(((((((((((.((((((((.......))))).))))))).))...)))))..))).)))------ ( -32.40, z-score = -1.00, R) >dp4.chr4_group4 2666485 137 + 6586962 UGGAGCCAAAGUGGCAUAUUUCUGAACCAACCUUCAGAUUUGUGUCUCAAAUACUCCGAUAAUAACACGAUAGACGAACGGCAUCCUGAAUAAUCCCUUAUAUUAUAACAGAUGACUUGUGUUCGUGUACAAUAGAC--- .((((.......((((((..((((((......))))))..)))))).......)))).......((((((.(.((((....((((.((.(((((.......)))))..))))))..)))).))))))).........--- ( -32.84, z-score = -2.33, R) >droPer1.super_10 1674683 137 + 3432795 UGGAGCUAAAGUGGCAUAUUUCUGAACAAACCUUCAGAUUUGUGUCUCAAAUACUCCAAUAAAAACUAGAUAGACGAACGGCAUCCUGAAUAAUCCCUCGUAUUAUAACAGACGACUUGUGUUCGUGUACAAUAGGC--- (((((.......((((((..((((((......))))))..)))))).......))))).......(((..((.((((((((.((........)).))((((..........)))).....)))))).))...)))..--- ( -32.74, z-score = -2.06, R) >droWil1.scaffold_180764 2077324 137 + 3949147 CGGAGCCAAAGUGGCAUAUUUAGUUUCCAUUCUUACAAUUUGUGUCUCAAAUGCUCCGAAAUCAAAUUACGAUCCGAACGGCAUCCUGUUUAAUCCUUCAUAUUAAAACAGAUGACUUAUGUUUGUGUACAAUC--UCC- ((((((....(.((((((..(.((..........)).)..)))))).)....)))))).........((((...((((((.((((.(((((..............))))))))).....)))))))))).....--...- ( -25.74, z-score = -0.90, R) >droGri2.scaffold_15252 2145982 132 - 17193109 UGGAGUCAAAGUGGCAUAUUUCAGAAUU-AUCCUCUGAUUUGUGUCUCAAAUGCUCCCAUAUUAA-UGAAUUCACGAACGGCAUUCUGUUCAAUAAUUUAUAUUGAUGCAGUU-ACUAGU-UUCGUGCACAGUUUC---- .(((((....(.((((((..(((((...-....)))))..)))))).)....)))))........-......(((((((......((((((((((.....)))))).))))..-....).-)))))).........---- ( -33.90, z-score = -2.67, R) >droVir3.scaffold_12963 2581484 138 - 20206255 UGGAGUUAAAGUGGCAUAUUUUGUGAAU-UUAACCGAAUUUGUGUCUCAAAUGCCCCAAAACUAA-CUAAAAGGCGAACGGCAUCCUGUUUAAUCCCUCGUAUUGAAACAGAUGACUUGUGUUUGUGUACAAUCGAUCCA .(((((((((.(.(((.....))).).)-)))))(((..(((((.(.((((((((..........-......)))..((((((((.(((((..............))))))))).).)))))))).))))))))).))). ( -26.23, z-score = 0.22, R) >droMoj3.scaffold_6500 9688105 138 - 32352404 UGGAGUCAAAGUGGCAUAUUCCGGGAUU-UCCUCUCGAUCUGUGUCUCAAAUACUCCCAAAU-AAACCUAAAGACGAACGGCAUCCUGUUUAAUUAAUCAUAUUAAUACAGAUGACUUGUGUUCGUGUACACCUAACCCA .(((((....(.((((((...(((((..-...)))))...)))))).)....))))).....-........(.((((((((((((.((((((((.......))))).))))))).))...)))))).)............ ( -30.40, z-score = -1.30, R) >consensus UGGAGUCAAAGUGGCAUAUUUUAGAACU_UUCCUCUGAUUUGUGUCUCAAAUGCUCCAAUAUCAACUUGAUAGACGAACGGCAUCCUGUUUAAUUACUAAUAUUAAAACAGAUGACUUGUGUUCGUGUACAACUUAA___ .(((((....(.((((((..(((............)))..)))))).)....)))))................(((((((.((..(((((((((.......))))).))))......))))))))).............. (-22.34 = -21.09 + -1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:14 2011